Figure 2.
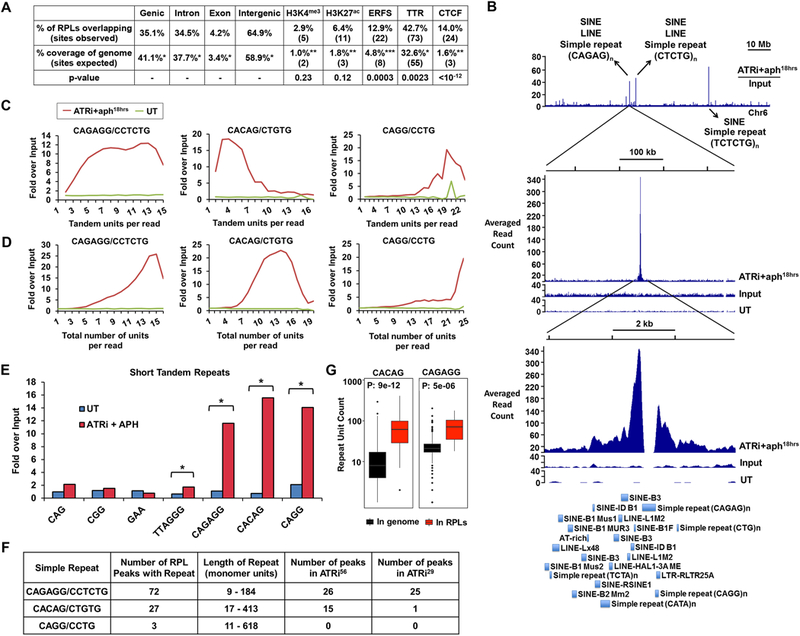
Short Tandem Repeats are Enriched in RPLs (A) Genomic features associated with RPLs. Percent and number of RPLs overlapping with noted features compared to expected overlap based on fraction of the genome comprised by these features is shown. Statistical significance (p value) was calculated by permutation test. (B) Example of repetitive DNA in RPL peaks. Top track: Representative ratio track of RPA-ChIP Seq reads over input reads from ATRi+aph18hrs-treated cells. Arrows detail examples of repetitive elements present. Middle and bottom track: Zoomed-in RPL peak. First track: RPA-ChIP of ATRi+aph18hrs; second track: input of ATRi+aph18hrs; third track: RPA-ChIP of DMSO-treated control (UT). Bottom: RepeatMasker annotations of repetitive elements within the peak region. (C, D) Quantification of tandem and total repeat units in RPA-ChIP Seq reads by REQer. X-axis depicts the (C) tandem repeat units and (D) total repeat units counted within the total RPA-ChIP Seq reads (ATRi+aph18hrs and DMSO control, UT) normalized by repeat occurrence in respective inputs. (E) Fold enrichment of tandem repeat occurrences in RPA-ChIP Seq NGS reads over input (average of 5 data points) in ATRi+aph18hrs and DMSO control (UT) is shown. *, p < 0.001, Student’s T-test. (F) Repeats most frequently observed as enriched in ATRi+aph18hrs RPA-ChIP Seq reads and their association with RPLs. (G) Lengths of CACAG and CAGAGG repeats in the mouse genome and in RPL peaks according to the reference genome. See also Figure S2 and S3.
