Figure 5.
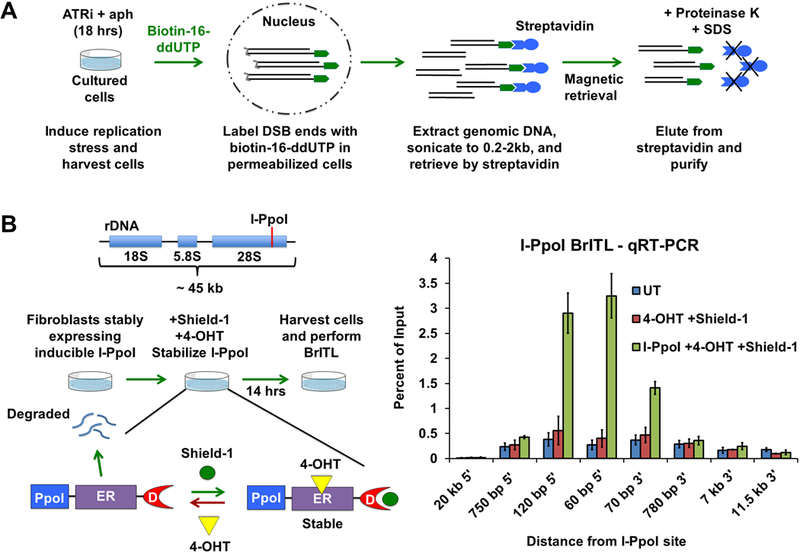
Development of BrITL (A) Schematic of the BrITL procedure. Treated cells are permeabilized and incubated with terminal deoxynucleotidyl transferase (TdT) and biotin-16-ddUTP. Extracted genomic DNA is then sonicated to 0.2–2 kb and subjected to streptavidin retrieval for analysis by qRT-PCR or NGS. (B) Validation of DSB detection by BrITL. Left: Genomic DSBs at I-PpoI recognition sites (red line) were conditionally generated by expression of I-PpoI (PpoI) fused to a destabilized FKBP12 (D) and a tamoxifen-specific form of the estrogen receptor (ER), followed by fusion protein stabilization and nuclear localization by Shield-1 and 4-hydroxytamoxifen (4-OHT) treatment. Right: qRT-PCR analysis of BrITL retrievals. Quantification of retrieved biotin-labeled fragments, normalized as a percent of input, is shown at specified distances from the rDNA I-PpoI site relative to the start of transcription. Conditions include UT (DMSO treatment), 4-OHT + Shield-1, and I-PpoI fusion expression + 4-OHT + Shield-1. For (B), the data are represented as mean +/− SEM.
