Abstract
Predicting 3D structure of protein from its amino acid sequence is one of the most important unsolved problems in biophysics and computational biology. This paper attempts to give a comprehensive introduction of the most recent effort and progress on protein structure prediction. Following the general flowchart of structure prediction, related concepts and methods are presented and discussed. Moreover, brief introductions are made to several widely-used prediction methods and the community-wide critical assessment of protein structure prediction (CASP) experiments.
Keywords: Protein structure prediction, homology modeling, ab initio prediction, structure refinement
1. Introduction
Since the latter half of 20th century, a growing number of researchers from diverse academic backgrounds are devoted to bio-related researches. Protein, as one of the most widespread and complicated macromolecules within life organisms, attracts a great deal of attentions. Proteins differ from one another primarily in their sequence of amino acids, which usually results in different spatial shape and structure and therefore different biological functionalities in cells. However, so for little is known about how protein folds into the specific three-dimensional structure from its one-dimensional sequence. In comparison with the genetic code by which a triple-nucleotide codon in a nucleic acid sequence specifies a single amino acid in protein sequence, the relationship between protein sequence and its steric structure is called the second genetic code (or folding code).1
The various advanced experimental techniques to determine protein sequence and structure have been developed in the last several decades. The number of proteins depositing in both UniProt2 and Protein Data Bank (PDB)3 keeps almost an exponential growth, especially in last two decades. It’s much easier to obtain protein sequences than to obtain their structures. With the development of advanced DNA sequencing technology, the sequences of proteins have been rapidly accumulated. The UniProt/TrEMBL database contains currently more than 85 million of protein sequences. On the structure side, X-ray crystallography and NMR spectroscopy are currently the two major experimental techniques for protein structure determination. Both of them are, however, time- and manpower-consuming, and have their own technical limitations for different protein targets. As of April 2017, the number of protein structures in PDB increases to ~ 120,000, which counts however only < 0.2% of the protein sequences in the UniProt.
The computational methods for predicting protein structure from its amino acid sequence spring up like mushrooms since the end of 20th century. The research article by Anfinsen in 19734 demonstrated that all the information a protein needs to fold properly is encoded in its amino acid sequence (called Anfinsen’s dogma). From the physical point of view, the amino acid sequence determines the basic molecular composition of the protein, and its native structure corresponds to the most stable conformation with the lowest free energy. Although we know that the protein folding process is governed by various physical laws, it is very hard to give such complicated macromolecule (including its interaction with surrounding solvent molecules) an accurate physical description. On the other hand, the huge amount of conformational space needed to search through is also a crucial issue remaining to be solved on the basis of the current computing power. Many different approaches have been proposed, including the employment of coarse-grained physical model and optimized conformational search algorithms, to address these issues.
There is a basic observation that similar sequences from the same evolutionary family often adopt similar protein structures, which forms the foundation of homology modeling. So far it is the most accurate way to predict protein structure by taking its homologous structure in PDB as template. With the rapid growth of PDB database, an increasing proportion of target proteins can be predicted via homology modeling. When no structure with obvious sequence similarity to the target protein can be found in PDB, it is still possible to find out proteins with structural similarity to the target protein.5 The method to identify template structures from the PDB is called threading or fold recognition,6–8 which actually matches the target sequence to homologous and distant-homologous structures based on some algorithm and take the best matches as structural template. The basic premise for threading to work is that protein structure is highly conservative in evolution and the number of unique structural folds are limited in nature.9,10
Both homology modeling (based on sequence comparison) and threading methods (based on fold-recognition) can be called template-based structure prediction methods.11 Unlike homology modeling and threading methods, ab initio method aims to build structure from the first principles of physics which does not rely on any previously solved structure. The development of ab initio method is also the exploration of the second genetic code. However, successful ab initio methods are very rare and there are still many problems and challenges waiting to be conquered. Currently Nearly all protein structure prediction methods make use of the structural information from previously solved structures to some extent. Therefore, “template-free”12 is often used for naming the methods that do not belong to the category of homology modeling and threading.
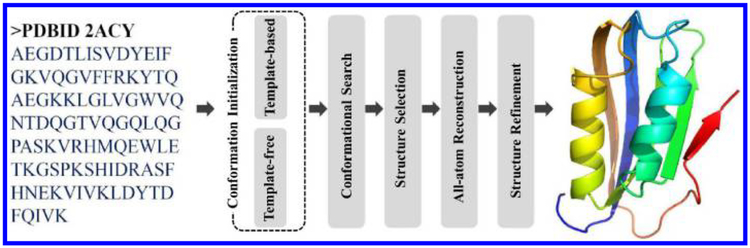
Although the detailed prediction processes involved in different prediction methods vary widely, the fundamental steps are consistent, including conformation initialization, conformational search, structure selection, all-atom structure reconstruction and structure refinement (as shown in Fig. 1). In this paper, we will introduce these steps in turn, including some well-known methods or tools commonly used in each step. We will also present a short overview of the critical assessment of protein structure prediction (CASP) experiments13,14 and some discussions about the current difficulties and future directions of protein structure prediction.
Fig. 1.
The general flowchart of protein structure prediction.
2. General Steps of Protein Structure Prediction
2.1. Conformation initialization
The starting point (input) of protein structure prediction is the one-dimensional amino acid sequence of target protein and the ending point (output) is the model of three-dimensional structures. The theoretically possible steric conformation for a protein sequence is almost infinite, but the native one for most protein is unique. It is very difficult to fold a protein from its amino acid sequence alone. First, we are still unable to construct a sufficiently accurate force field that can guide the target sequence folding in the right direction; second, the amount of computation involved in such a vast conformational search process can easily go beyond the existing computing ability.
Actually, the key difference between the “template-based” and “template-free” methods is the way of conformation initialization. The template-based method obtains the initial conformation by searching for the solved structures which are homologous or structurally similar with the target protein. The template-free method usually constructs the initial conformation by fragment assembly. As of now, most of the successful modeling methods, including the template-free methods, somehow use the structural information from PDB database, by which an initial conformation much better than random ones can be quickly built. Thus the computing load for conformational search can be significantly reduced and the reliability of the prediction can be greatly enhanced as well.
In most cases the structural template homologous to the target protein can be identified from the PDB database by sequence alignment,15–17 and an accurate alignment between target and template can be built. The similarity between sequences often implies the similarity between their structures. Sometimes the conformation copied from the template (not necessarily complete) is already very close to the optimal one; this is especially the case to homology modeling, where the key issue for homology modeling is thus to improve the efficiency and accuracy of sequence alignment. Since the protein structure is much more conservative than its sequence, it is often the case that two proteins without any sequential similarity fold into similar folds, which is actually the foundation of threading methods. During the past two decades, the threading method has been widely studied and applied, which greatly improves the utilization of the experimentally solved structures. In general, the initial conformation obtained from structural template is vastly better than any ones built from scratch and can dramatically shorten the process of subsequent conformational search.
However, there is no guarantee that the satisfactory structural templates for any target protein can always be found. The template-free methods are the best choice for the hard target proteins of which no satisfactory template can be identified. It is the most straightforward way to generate the initial conformation of target protein by random; but in this way the burden of conformational search would be very heavy. Along with the inadequacy of current force field, it’s extremely difficult to accomplish the simulation process with such huge conformational change. In fact, the complex and multilevel nature of protein structure provides us with more choices. We can first predict the specific structural features of target protein, such as backbone dihedral angle, solvent accessibility, contact map or secondary structure, which does not necessarily depend on any structural template. Many template-free methods18,19 predict protein structure by fragment assembly, where the corresponding structural fragments are usually cut from experimental structures based on the prediction of secondary structure, backbone dihedral angle and so on. The conformation built by fragment assembly would be dramatically better than any random conformation, and can especially guarantee better local structure quality.
2.2. Conformational search
After the initial conformation is constructed, we can continue to run simulation with the guide of a certain force field to search for near-native conformations step by step. As a typical biological macromolecule, protein consists of thousands of atoms and its conformational degrees of freedom are huge. Therefore, a simplified representation of protein conformations becomes particularly crucial for speeding up the simulation of protein folding process. In fact, the structural template identified by sequence alignment is already a reduced conformation with only backbone or Cα-atoms, because sequence alignment is actually in residue-level and the matches of different residues make the side chain conformations from template unusable to target protein. Currently almost all protein structure assembly simulation methods do conformational search based on a certain kind of simplified representation. For example, each residue can be represented only by its Cα-atom and the virtual center of side chain, or the entire backbone conformation can be represented by a series of dihedral angles.
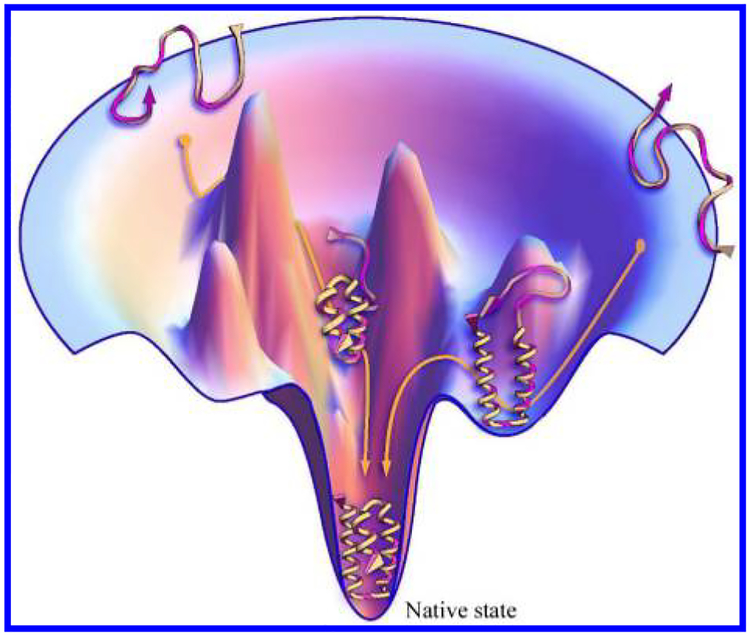
A force field that can depict the protein conformational energy landscape is needed for conformational search. Any conformation of the target protein corresponds to a point on the energy landscape. For a well-designed force field, the native conformation of the target protein should be at the lowest point of the energy landscape (as shown in Fig. 2). Since the protein folding process results from the atomic interactions, the best description of that process should be based on quantum mechanical theory. However, so far only very small molecular systems can be calculated based on quantum mechanical theory due to the limited computing capability. A full quantum mechanical treatment for a complete protein is not feasible.
Fig. 2.
Protein folding guided by funnel-shaped energy landscape (see Ref. 20).
Many existing force fields are simply based on the classical physics theory, which generally consists of the bond-stretching energy, the angle bending energy, the angle torsional energy and other energies for nonbonded interactions. These types of force fields are widely used in molecular dynamics simulation, like AMBER21 and CHARMM.22 They are also referred to as physics-based energy function. In contrast, the other type of energy function is constructed purely based on the information of structural features derived from the experimentally solved structures, and therefore they are usually referred to knowledge-based energy function.23,24 The PDB actually serves as a vast treasure trove for protein structure prediction. It is not only a structural template library for homology modeling, but also a reliable structural information pool for designing knowledge-based energy function. In 1990, Sippl25 proposed a statistical potential based on the distance distribution of residue pair and used it to evaluate the backbone conformation of peptide. Subsequently, many distance-dependent atom-pair potentials26–29 were developed based on the similar ideas to Sippl, which differ from each other mainly in their reference states.30 In addition to distance, other structural features from native structure can be also used to construct knowledge-based energy function, such as dihedral angles, solvent accessibility, side chain orientation and so on. Various machine learning methods (e.g., hidden Markov model, artificial neural network and support vector machine) have been used for deriving such knowledge-based energy function.
These two types of energy functions have their own advantages and disadvantages respectively. The physics-based energy functions are constructed from the first principle and have a clear physical significance, but they are often inaccurate for describing the complicated atomic interactions and have poor performances in protein structure prediction. The knowledge-based energy functions make full use of experimentally solved structures and exhibit promising performance in many cases, but they are like “black boxes” which cannot help us understand the nature of protein folding process. In fact, most of structure prediction methods employ both physics-based energy function and knowledge-based energy function.
Once the energy function is determined, we need to find a proper way to search for the lowest-energy conformation of target protein. Molecular dynamics simulation31,32 is commonly used for conformational search, which simulate protein folding and movement by solving Newtonian motion equations. However, as for biomacromolecule like protein, the demand of molecular dynamics simulation for computing resources is too big. In general, the time step of molecular dynamics simulation is in the order of femtosecond (10−15 s), while the time required for protein folding is usually in milliseconds (10−3 s). So for it is still a great challenge to fold a protein by molecular dynamics simulation. Nevertheless, the conformational search process based on Monte Carlo simulation33,34 can be much faster, which randomly changes the conformation based on various movements designed beforehand. These movements include the shifts and rotations of structural segment as well as the position changes of a single atom. They can be spatially continuous, or confined to a cubic lattice system (which can dramatically reduce the conformation space). Moreover, many strategies, such as simulated annealing35 and replica exchange,36,37 are widely used to help cross over the energy barriers and achieve the global minimum energy state.
2.3. Structure selection
Following the conformational search, a large number of structures of target protein are generated. One of the unsolved issues in both molecular dynamics simulation and Monte Carlo simulation is that the conformations are often trapped at the local minimal state. Even with the global minimal state identified, the conformation is not necessarily corresponding to the one closest to native state because of the inadequacies of force field. Thus, the common procedure during simulation is to regularly output lower energy intermediate structures for subsequent conformational screening. The key factor of structure selection is the assessment method for distinguishing native-like structures from nonnative ones. There is a specific prediction category in CASP for assessing the methods of structural quality assessment.38 It should be noted that the methods for structure selection may be designed specifically for assessing the reduced structural models corresponding to the simplified representation adopted during conformational search. It is an important research direction in protein structure prediction to develop methods of structural quality assessment based on all kinds of ideas and techniques. Furthermore, there are many structural decoys generated by diverse methods39–41 available for training and testing methods of structural quality assessment, including the structural models from CASP.
In fact, the force field itself can filter structures, and the output of conformational search is just the filtered structures by the corresponding force field. But what needs to be carefully considered in designing force field is the contradiction between the accuracy and the speed. Since the number of structures for selection is dramatically less than that the force field of conformational search needs to deal with, we can use a much more complicated energy function for structure selection. The energy function can be either physics-based or knowledge-based, while the latter is more popular and effective. Moreover, there are other ways to select structure by clustering all structures based on structure similarity.42–44 It is generally believed that the structure with a higher frequency of occurrence has a lower free energy, which is the theoretical basis of structural clustering methods.
2.4. All-atom structure reconstruction
Since most of prediction methods adopt simplified protein representation for conformational search, so far what we have obtained are just one or several reduced structural models. The all-atom structure should be reconstructed based on the reduced models. The process of all-atom reconstruction varies a lot for reduced models based on different protein representation. Some prediction methods adopt the representation of “Cα atom” plus “virtual center of side chain”, where the “virtual center of side chain” only acts as an assistant for determining the position of Cα atom during conformational search and the output structure contains only Cα atoms. In that case, the reconstruction process is usually divided into two separate steps. The first step is to rebuild the backbone atoms (C N and O) based on the position of Cα atoms, which is the primary function of many methods developed specifically for all-atom reconstruction, such as SABBAC,45 BBQ,46 PULCHRA47 and REMO.48 All these methods depend on the backbone fragments cut from experimental structures. For example, the backbone isomer library built by REMO contains 528798 fragments with four consecutive residues which are collected from 2561 protein chains in PDB. The second step is to rebuild the side chain for every residue. Like the strategy used in rebuilding backbone atoms, the best way to rebuild side chain is also based on the related library (side-chain ro-tamer library).49 There are many methods for side chain reconstruction too, such as Scwrl,50,51 SCATD,52 RASP,53 and so on.
2.5. Structure refinement
Although the complete structure of the target protein has been obtained by the previous steps, the structural quality is usually not very good, which may owe to the defects of the force field, conformational search or all-atom reconstruction. The process of structure selection by clustering method may also bring some local structural issues if the structures of cluster centroid are used.54 Therefore, it is almost a routine step to further refine the structure after all-atom reconstruction. Since the structural issues in reduced model can directly affect the quality of final allatom structure, some methods combine the procedures of all-atom reconstruction and refinement.55 They refine the reduced model (such as backbone structure) and all-atom structure separately according to the reconstruction schedule.
Structure refinement also requires a force field to conduct molecular dynamics simulation or Monte Carlo simulation, but this procedure is quite different from the previous step of conformational search. The aim of conformational search in structure assembly simulations is to determine the backbone structure of the target protein, which actually sacrifices the structural details to ensure the search efficiency. However, the main purpose of structure refinement is to improve the quality of allatom structure (especially local structure) where only small change is conducted in backbone conformation. It is highly nontrivial to refine the overall topology as well as the structural details of the target protein. The conformational changes involved in structure refinement are much smaller than those in structure assembly simulations, which offers a feasible task for molecular dynamics simulation even in all-atom level. FG-MD56 is one of such methods for structure refinement based on atomic-level molecular dynamics simulation, which collects spatial restraints from both global templates and local fragments to guide the structure refinement. Mod-Refiner55 is another method for structure reconstruction and refinement based on Monte Carlo simulation. It can first reconstruct the backbone structure from initial Ca trace and conduct energy minimization simulation to refine the backbone quality, then add the side chain atoms and conduct another round of simulation for minimizing the energy of all-atom structure. A prediction category for model refinement is introduced since CASP7 in order to assess the abilities and inabilities in the area of structure refinement.57–59
3. Two Categories of Protein Structure Prediction Methods
Although there are a series of important steps for predicting protein structure and each step belongs to an independent research field where lots of related methods have been developed, what the users of the structure prediction methods usually concern are ease-of-use, efficiency and reliability of the prediction method, not the specific prediction steps and the related techniques. After decades of developments, there are many distinguished prediction methods being built and available for users around the world.60,61 In this section, we will give a brief introduction to several widely-used prediction methods, including both template-based and template-free ones.
3.1. Template-based methods
For most target proteins, the desirable structural template can be identified from PDB by sequence alignment or threading method. Since the conformational information from template is much more reliable than that from elsewhere (especially when the target protein and the template are highly homologous), the prediction accuracy of template-based method is generally higher than other methods, which makes it highly popular in practical applications.
SWISS-MODEL62,63 is a well-known online tool developed by Torsten Schwede’s structural bioinformatics group, which is dedicated to homology modeling of protein structures. The prediction process consists of template recognition, target-template alignment, model building and model evaluation. BLAST17 and HHblits64 are employed for template recognition and target-template alignment. The structure of the target protein is built by copying the atomic coordinates from the template according to target-template alignment. The unaligned region is constructed by searching the fragment library. Final model are evaluated by the knowledge-based score function QMEAN.65 There are three types of modeling modes: automated mode, alignment mode and project mode, which differ mainly in the amount of user intervention. It also provides the module for oligomeric structure prediction. The rich functionality, easy operability and fast running make SWISS-MODEL one of the most widely-used homology modeling tool in the past two decades. For information about the accuracy, stability and reliability of the SWISS-MODEL, the reader can visit CAMEO website (https://www.cameo3d.org/)66 which provide continuous evaluation of protein structure prediction services in a fully automated manner.
Modeller67,68 is another homologous modeling method developed by the Andrej Sali Lab, which implements structure modeling by satisfaction of spatial restraints. The spatial restraints can be derived from the target-template alignment and many other sources, such as data of NMR spectroscopy, fluorescence spectroscopy, site-directed mutagenesis, stereochemistry and intuition. The types of spatial restraints include bond lengths, bond angles, dihedral angles, non-bonded atom-atom contacts and so on. It is very convenient for Modeller to make use of all kinds of information by converting them into spatial restraints. The final model will be evaluated by DOPE potential.69 Modeller can also perform additional auxiliary tasks, including fold assignment, calculation of phylogenetic trees and de novo modeling of loops in protein structures. The executable version of the Modeller program is available for download and installation on the local computers of various systems. Due to Modeller’s popularity, there are several third party GUIs for Modeller being developed in recent years.70,71
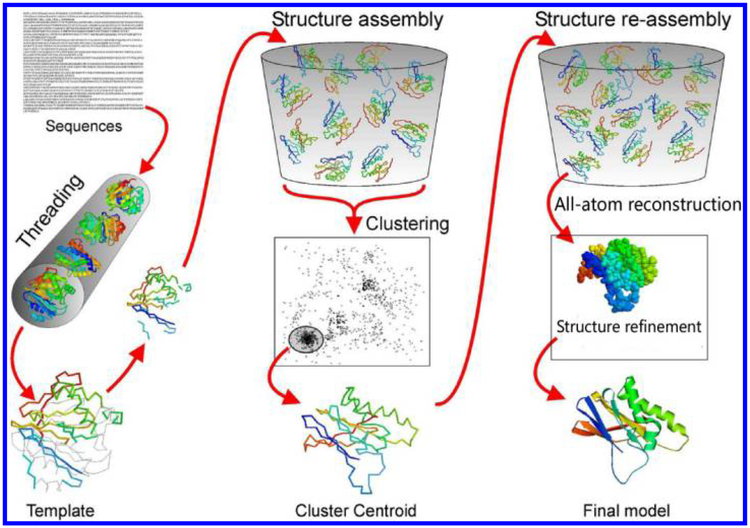
Homology modeling usually requires that the target and the template share notable sequence identity (e.g. > 25%). Protein threading can go beyond this limitation and achieve a better performance even when the sequence identity is very low. I-TASSER72,73 is a comprehensive structure prediction method based on threading method developed by the Yang Zhang Lab. The flowchart of I-TASSER is shown in Fig. 3. It first identifies structural templates or super-secondary structure fragments from a non-redundant subset of PDB by multiple threading approach LOMETS.74 Second, with the initial conformations built from the templates, a large number of reduced models are generated by replica-exchange Monte Carlo simulations. Third, all reduced models are clustered by SPICKER43 and the cluster centroid are created by averaging the coordinates of all decoys in each cluster. Fourth, the fragment assembly simulation is performed again starting from the selected cluster centroids. Fifth, all-atom structures are reconstructed and refined by FG-MD.56 Finally, up to five full-length atomic models along with the estimated accuracy of the models are outputted. As a comprehensive method, I-TASSER performs pretty well even for new fold targets. It keeps ahead in the last decade of the community-wide CASP experiments. Users can submit their target sequence to I-TASSER webserver or download the package of I-TASSER Suite for running on their local computers.
Fig. 3.
Flowchart of I-TASSER method for protein structure prediction.
3.2. Template-free methods
Currently most structure prediction methods rely on the information provided by the experimental structures (the most direct way is the use of structural templates), which is not helpful for us to explore and understand the essential law of protein folding. The development of template-free methods is driven not only by the practical application (not all target proteins can find a satisfactory template in PDB), but also by the basic scientific problem of protein folding code. Although the template-free methods commonly exploit the information from known structures as well, their development can better reflect the theoretical and technical level of protein structure prediction than template-based methods.
Rosetta18,75 is a template-free method developed by the David Baker Lab, which assembles full-length structure based on fragments of 3–9 residues from PDB structures. Similar to template-based methods, these fragments are selected on the basis of local sequence similarity and the similarity between the known and predicted secondary structure. The assembly simulation is then conducted by Monte Carlo simulated annealing search strategy. QUARK19 is another excellent fragment-assembly method developed by the Yang Zhang Lab. The structural fragments used by QUARK range from 1 to 20 residues, with the assembly simulation conducted by the replica-exchange Monte Carlo simulation under the guide of an atomic-level knowledge-based force field. There are many other methods which are also based on fragment assembly, such as SCRATCH,76 PROFESY,77 FRAGFOLD,78 and so on. The key difference between these methods and the template-based methods lies in that they do not rely on any global structural template and require no homology or structure similarity between the target protein and the proteins where the fragments come from. Therefore, it can be more capable for template-free methods to model target of new folds. However, it is still a great challenge for template-free methods to model proteins with length > 150 residues because of the huge computing demand and low accuracy of force field. Contact map prediction based on co-evolution approach recently demonstrated promise in breaking up such length limit of ab initio structure folding.
4. The Critical Assessment of Protein Structure Prediction Experiments
When talking about protein structure prediction, one important topic that cannot be bypassed is the CASP experiments.13,60 The first CASP experiment was launched in 1994 by John Moult at the University of Maryland. After that, the CASP experiment is held every two years with the latest being CASP12 in 2016 at the time of preparing this review. CASP provides the participants with a worldwide platform to objectively evaluate their prediction methods and to obtain an overall comparison with other methods. More importantly, the CASP experiment can help establish the current state of the art in protein structure prediction, demonstrate what progress has been made, and highlight where future effort may be most productively focused.
The targets for structure prediction are selected by the organizers who are not permitted to participate in the CASP experiment. These target proteins are either not experimentally solved yet or already solved but not yet publicly accessible. Depending on whether a suitable template can be identified or not, the domains of all target proteins are divided into two basic categories, the template based modeling (TBM) and the template-free modeling (FM). Then pertinent assessments can be implemented on the predicted models of each category. Furthermore, prediction methods are also divided into two categories, those using a combination of computational methods and human experience (Human Section), and those relying solely on computational methods (Server Section). Generally, a window of three weeks is provided for prediction of a target by human-expert groups and three days by servers. After the closing of the server prediction window, the server models are posted at the Prediction Center web site (http://predictioncenter.org), which can be further used by human-expert predictors as starting points for more detailed modeling. Once all models of a target protein have been collected, the Prediction Center performs a standard numerical evaluation of the models by taking the experimental structure as the gold standard. The identity of the predictors is concealed from the assessors when they conduct their analysis. At the predictors’ meeting in December of a CASP year, the results of the evaluations are presented to the community and also made publicly available through the Prediction Center web site. The articles by the organizers, the assessors, and the most successful prediction groups are published in special issues of the journal Proteins: Structure, Function, and Bioinformatics.
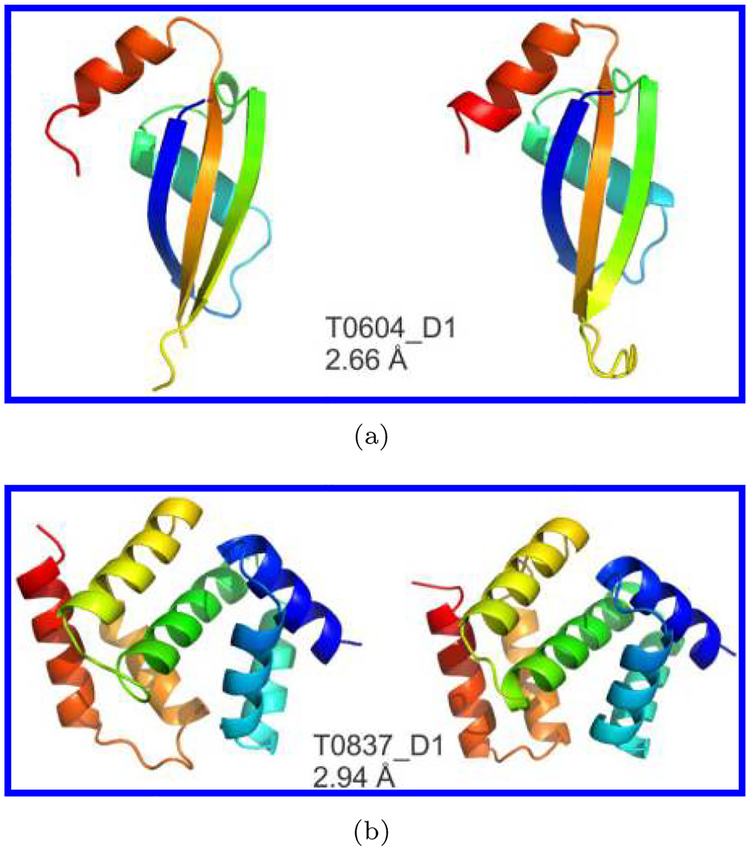
The CASP experiments have witnessed the history of the development of protein structure prediction over the past two decades. In the first few CASP experiments, the homology models are generally even farther away from the target than the initial template. But in recent CASP experiments, the best models of many target proteins are moved much closer to the target than the best templates, which would mainly benefit from the development of multiple-template modeling and the improvement of force fields. The template-free methods have also made great progress. For the FM targets with about 100 residues, many template-free methods can build models with correct global topology (RMSD to native is about 4–10 Å, in some cases close to 2 Å). Figure 4 shows two successful examples from CASP experiments. The first one is the first model of T0604_D1 in CASP9 by I-TASSER, which benefits much from sequence-based contact prediction, not from sequence alignment. The second one is the first model of T0837_D1 in CASP11 by QUARK, which also effectively adopted the contact information derived from fragments. Most recently, it has been found in CASP12 that some FM targets with a larger size can be successfully folded with the assistance of the coevolution based contact map prediction. However, the success relies on the availability of the high number of sequence homologous sequences so that reliably coevolution information can be derived from the high-dimension multiple sequence alignment matrix. Thus, it remains a great challenge for template-free methods to obtain highly accurate models for target proteins larger than 150 residues, when the coevolution based contact-map information is not available.
Fig. 4.
Two examples of ab initio modeling in CASP (the left and right panels are X-ray structures and predicted models respectively). (a) The first model of T0604_D1 in CASP9 by I-TASSER, with RMSD = 2.66 Å, length = 79, and classification being FM target; (b) the first model of T0837_D1 in CASP11 by QUARK, with RMSD = 2.94 Å, length = 128, and classification being FM target.
One of the most important goals of the CASP experiments is to promote the development of template-free methods. Due to the rapid growth of experimental structures in the PDB, the protein sequences that can be classified as the FM targets are becoming increasingly scarce. Therefore, since 2011, the organizers introduced a new prediction category called CASP ROLL,79 aiming to collect FM targets outside the regular CASP season for the same double-blind prediction and assessment as the regular CASP tertiary structure evaluation.
Besides the tertiary protein structure prediction, CASP evaluates several other structure-related modeling categories, such as residue-residue contacts,80 disorder region prediction,81 model quality assessment (MQA) methods,38 tertiary structure refinement,58 and so on. All models and assessment results of the previous CASP experiments are publicly available at the Prediction Center web site, allowing predictors to compare their own models with those submitted by other groups. Readers can also refer to the publications of previous experiments (http://predictioncenter.org/index.cgi?page=proceedings) for more detailed information.
5. Summary and Prospect
This review gives a brief introduction of protein structure prediction, including the background, general steps, representative methods and the CASP experiments. We hope it can be helpful for readers to obtain an overall picture of protein structure prediction. Nowadays, protein structure prediction is one of the most representative and influential research areas in computational biology and bioinformatics; it is however not an isolated scientific problem, but a comprehensive field involving multidisciplines. The development of protein structure prediction is closely related to the improvements of alignment search algorithm, data mining technology, energy function design and molecular simulation. Up to now, many prediction methods are already quite mature and are widely used by molecular biologists all over the world.82 For instance, the I-TASSER online servers (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) have provided protein structure prediction services for more than 80 thousand researchers from 130 countries and regions during the last decade.
Nevertheless, there are still many problems and challenges to overcome, for example: (i) the performances of current alignment/threading algorithms for remote homology templates remain to be improved; (ii) the force field for conformational search is not accurate enough, or not the optimal to the specific protein representation; (iii) the global topology and the local structure are difficult to be refined simultaneously; (iv) the membrane proteins, whose structures are difficult to determine by experiment, are also hard nuts for computational prediction; (v) the prediction of protein complex (quaternary structure) is of great importance but currently is far from satisfactory. We believe that all these difficulties will eventually be resolved with the development of the study. Moreover, as the organizers of CASP experiments have pointed out, currently most of the prediction methods (including template-free methods) rely too much on the known structural information, which is what we do not want to see. The new breakthrough in protein structure prediction can be expected by reducing reliance on known structures and enhancing first-principles research. We look forward to the unraveling of the second genetic code by protein structure prediction in the foreseeable future.
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities (Grant No. 2662015BQ045), the National Natural Science Foundation of China (Grant Nos. 11604111 and 11474117) and the National Institute of General Medical Sciences (GM083107 and GM116960).
References
- 1.Kolata G, Science 233, 1037 (1986). [DOI] [PubMed] [Google Scholar]
- 2.Consortium U, Nucl. Acids Res 43, D204 (2015).25348405 [Google Scholar]
- 3.Berman HM et al. , Nucl. Acids. Res 28, 235 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anfinsen CB, Science 181, 223 (1973). [DOI] [PubMed] [Google Scholar]
- 5.Bowie JU, Luthy R and Eisenberg D, Science 253, 164 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Jones DT, Taylor WR and Thornton JM, Nature 358, 86 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Jones D and Thornton J, J. Comput.-aided Mol. Des 7, 439 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Jones DT, J. Mol. Biol 287, 797 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Chothia C, Nature 357, 543 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y and Skolnick J, Nucl. Acids Res 33, 2302 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang YJP et al. , Protein Struct. Funct. Bioinf 82, 43 (2014). [Google Scholar]
- 12.Tai CH et al. , Protein, Struct. Func. Bioinf 82, 57 (2014). [Google Scholar]
- 13.Moult J, Curr. Opin. Struct. Biol 15, 285 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Kryshtafovych A, Fidelis K and Moult J, Intro. Protein Structure Prediction, Methods Algorithms (John Wiley and Sons, Inc., Hoboken, New Jersey, 2010), p. 15–32. [Google Scholar]
- 15.Needleman SB and Wunsch CD, J. Mol. Biol 48, 443 (1970). [DOI] [PubMed] [Google Scholar]
- 16.Smith TF and Waterman MS, J. Mol. Biol 147, 195 (1981). [DOI] [PubMed] [Google Scholar]
- 17.Altschul SF et al. , Nucl. Acids Res 25, 3389 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohl CA et al. , Methods Enzymol 383, 66 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Xu D and Zhang Y, Proteins, Struct. Funct. Bioinf 80, 1715 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dill KA and MacCallum JL, Science 338, 1042 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Pearlman DA et al. , Comput. Phys. Commun 91, 1 (1995). [Google Scholar]
- 22.Brooks BR et al. , J Comput Chem 4, 187 (1983). [Google Scholar]
- 23.Tanaka S and Scheraga HA, Macromolecules 9, 945 (1976). [DOI] [PubMed] [Google Scholar]
- 24.Miyazawa S and Jernigan RL, Macromolecules 18, 534 (1984). [Google Scholar]
- 25.Sippl MJ, J. Micromol. Biol 213, 859 (1990). [DOI] [PubMed] [Google Scholar]
- 26.Samudrala R and Moult J, J. Mol. Biol 275, 895 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Lu H and Skolnick J, Proteins, Struct. Funct. Bioinf 44, 223 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Zhou H and Zhou Y, Protein Sci 11, 2714 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rykunov D and Fischer A, BMC Bioinf 11, 128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng H et al. , Proteins, Struct. Funct. Bioinf 80, 2311 (2012). [Google Scholar]
- 31.Van Gunsteren WF et al. , Angew. Chem. Int. Edn 45, 4064 (2006). [Google Scholar]
- 32.Sugita Y and Okamoto Y, Chem. Phys. Lett 314, 141 (1999). [Google Scholar]
- 33.Hansmann UHE and Okamoto Y, Curr. Opin. Struct. Biol 9, 177 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Li Z and Scheraga HA, Proc. Natl. Acad. Sci. USA 84, 6611 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirkpatrick SC, Gelatt CD and Vecchi MP, Science 220, 671 (1983). [DOI] [PubMed] [Google Scholar]
- 36.Swendsen RH and Wang JS, Phys. Rev. Lett 57, 2607 (1986). [DOI] [PubMed] [Google Scholar]
- 37.Kihara D et al. , Proc. Natl. Acad. Sci 98, 10125 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kryshtafovych A et al. , Proteins, Struct. Funct. Bioinf 82, 112 (2014). [Google Scholar]
- 39.Samudrala R and Levitt M, Protein Sci 9, 1399 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai J et al. , Proteins, Struct. Funct. Genetics 53, 76 (2003). [Google Scholar]
- 41.Deng H, Jia Y and Zhang Y, Bioinformatics 32, 378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shortle D, Simons KT and Baker D, Proc. Natl. Acad. Sci. USA 95, 11158 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y and Skolnick J, J. Comput. Chem 25, 865 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Kozakov D et al. , Biophys. J 89, 867 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maupetit J, Gautier R and Tuffery P, Nucl. Acids Res 34, W147 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gront D, Kmiecik S and Kolinski A, J. Comput. Chem 28, 1593 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Rotkiewicz P and Skolnick J, J. Comput. Chem 29, 1460 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li YQ and Zhang Y, Proteins Struct. Funct. Bioinform 76, 665 (2009). [Google Scholar]
- 49.Dunbrack RL and Karplus M, J. Mol. Biol 230, 543 (1993). [DOI] [PubMed] [Google Scholar]
- 50.Krivov GG, Shapovalov MV and Dunbrack RL, Proteins, Struct. Funct. Bioinform 77, 778 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Canutescu AA, Shelenkov AA and Dunbrack RL, Protein Sci 12, 2001 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu J, Rapid Protein Side-Chain Packing via Tree Decomposition (Springer-Verlag; Berlin Heidelberg, 2005), p. 423–439. [Google Scholar]
- 53.Miao Z, Cao Y and Jiang T, Bioinformatics 27, 3117 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Wu S, Skolnick J and Zhang Y, BMC Biol 5, 17 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu D and Zhang Y, Biophys. J 101, 2525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Liang Y and Zhang Y, Structure 19, 1784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacCallum JL et al. , Proteins, Struct. Funct. Bioinf 79, 74 (2011). [Google Scholar]
- 58.Nugent T, Cozzetto D and Jones DT, Proteins, Struct. Funct. Bioinf 82, 98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Modi V, Dunbrack RL Jr., Proteins 84(1), 260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moult J et al. , Proteins, Struct. Funct. Bioinform 84(S1), 4 (2016) [Google Scholar]
- 61.Moult J et al. , Proteins, Struct. Funct. Bioinform 82, 1 (2014). [Google Scholar]
- 62.Guex N and Peitsch MC, Electrophoresis 18, 2714 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Biasini M et al. , Nucl. Acids Res 42, 252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Remmert M et al. , Nature Methods 9, 173 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Benkert P, Kunzli M and Schwede T, Nucl. Acids Res 37, W510 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haas J et al. , Database J. Biol. Databases Curation 2013: bat031–bat031. [Google Scholar]
- 67.Sali A and Blundell TL, J. Mol. Biol 234, 779 (1993). [DOI] [PubMed] [Google Scholar]
- 68.Webb B and Sali A, Curr. Protoc. Bioinform 47, 1 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Shen MY and Sali A, Protein Sci 15, 2507 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuntal BK, Aparoy P and Reddanna P, BMC Res. Notes 3, 1 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathur A, Shankaracharya and Vidyarthi AS, J. Mol. Model 17, 2601 (2011). [DOI] [PubMed] [Google Scholar]
- 72.Roy A, Kucukural A and Zhang Y, Nat. Protoc 5, 725 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J et al. , Nature Methods 12, 7 (2014). [Google Scholar]
- 74.Wu S and Zhang Y, Nucl. Acids Res 35, 3375 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simons KT et al. , J. Mol. Biol 268, 209 (1997). [DOI] [PubMed] [Google Scholar]
- 76.Cheng J et al. , Nucl. Acids Res 33, 72 (2005). [Google Scholar]
- 77.Lee J et al. , Proteins Struct. Funct. Bioinform 56, 704 (2004). [Google Scholar]
- 78.Jones DT, Proteins Struct. Funct. Bioinform 5, 127 (2001). [Google Scholar]
- 79.Kryshtafovych A, Monastyrskyy B and Fidelis K, Proteins, Struct. Funct. Bioinf 82, 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Monastyrskyy B et al. , Proteins, Struct. Funct. Bioinform 82, 138 (2014). [Google Scholar]
- 81.Monastyrskyy B et al. , Proteins, Structure, Funct. Bioinf 82, 127 (2014). [Google Scholar]
- 82.Zhang Y, Curr. Opin. Struct. Biol 19, 145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]