Figure 1. Mitochondrial mutations are stably propagated in human cells in vitro.
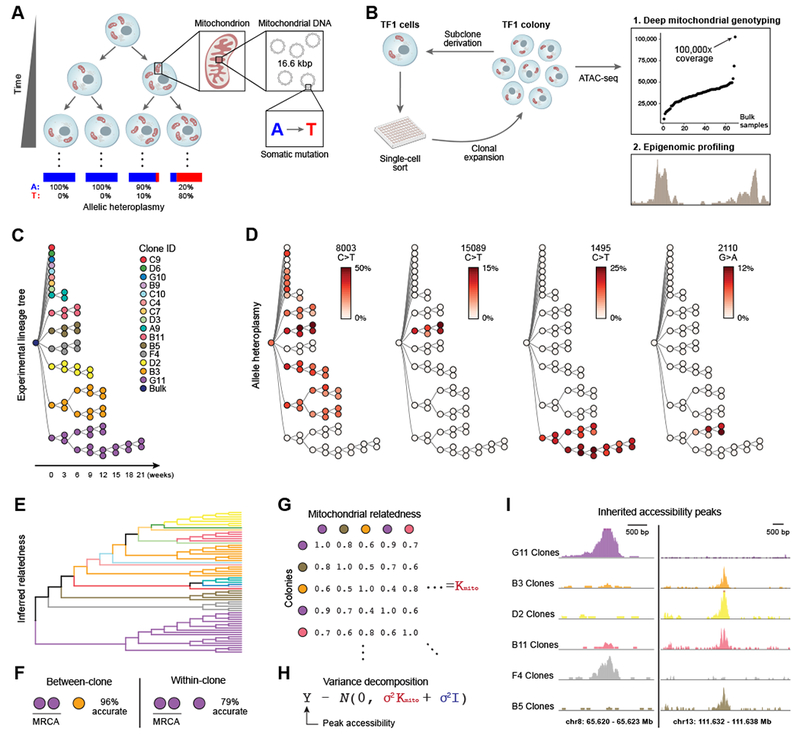
(A) Dynamics of mtDNA heteroplasmy in single cells. Each cell has multiple mitochondria, which in turn contain many copies of mtDNA that may acquire somatic mutations over time. (B) Proof-of-principle design. Each TF1 cell clone and sub-clone is assayed with ATAC-seq. (C) Supervised (true) experimental TF1 lineage tree. Colors indicate each primary clone from initial split. (D) Allelic heteroplasmy of four selected variants reveals stable propagation and clone-specificity. Color bar: allelic heteroplasmy (%). (E) Unsupervised hierarchical clustering of TF1 clones. Color: primary clones as in (C). (F) Between-clone and within-clone accuracy of identifying the most-recent common ancestor (MRCA) per trio of clones based on mtDNA mutational profile. (G) Schematic of mitochondrial relatedness matrix Kmito where each pair of clones is scored based on mitochondrial genotype similarity. (H) Random effects model for variance decomposition of epigenomic peaks. (I) Two examples of peaks inherited in clonal lineages. Peaks represent the sum of open chromatin for the clones with the most samples.
