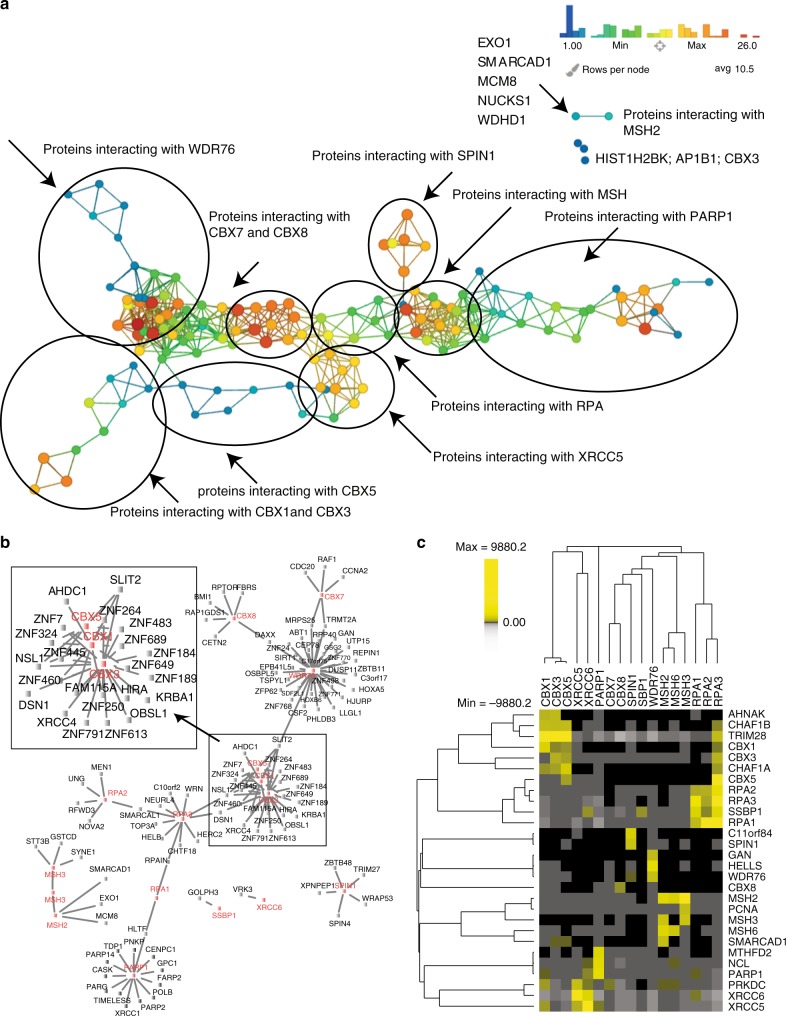
Fig. 3.
Topological network and linkage analysis of a human DNA repair network. a TDA in combination with the TopS scores was applied to the proteins with Tops > 20 in at least one of the baits. Norm Correlation was used as a distance metric with two filter functions: Neighborhood lens 1 and Neighborhood lens 2. Resolution 30 and gain 3 were used here using the Ayasdi software24. Proteins are colored based on the rows per node. Color bar: red: high values, blue: low values. Node size is proportional to the number of proteins in the node. b A reduced network was created using information from the CRAPome27 database, and the network was generated using Cytoscape platform23. Proteins associated with CBX1, CBX3, and CBX5 are expanded and highlighted in the inset. c Hierarchical biclustering using PermutMatrix47 of protein with extreme positive TopS values. The 28 proteins with the highest TopS values across the dataset are shown. Rows and columns were clustered using Pearson as the distance and Ward linkage as the method. Yellow corresponds to high TopS values and gray shows negative TopS values. Proteins not present in the purifications are shown in black. TopS topological scoring