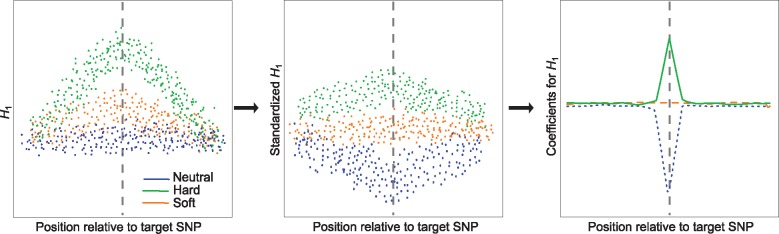
Fig. 1.
Schematic illustrating steps taken by Trendsetter to learn a multinomial regression model. For a given summary statistic (e.g., expected haplotype homozygosity H1), we compute its value spatially across a genomic region for a set of neutral, hard sweep, and soft sweep simulations used as training data. For H1, we expect elevated values near the site under selection (target SNP; indicated by a gray vertical dashed line) in sweep simulations, and a greater magnitude of elevation in hard sweep compared with soft sweep settings. This summary statistic is then standardized (mean centered and normalized by the standard deviation) at each position it is computed, so that different summary statistics are comparable. For H1, this standardization will yield strong negative values for neutral simulations and positive values for hard sweep simulations near a target SNP, and soft sweep simulations will exhibit values intermediate between the neutral and hard sweep scenarios. The model then performs trend filtering on the spatial distribution of each summary statistic (here H1) for each class (here neutral, soft sweep, and hard sweep), leading to a curve describing the spatial distribution of summary statistics around a target SNP. For H1, the curve dramatically reduces for the neutral class near the center of the sequence, and is elevated near this position for the hard sweep class.