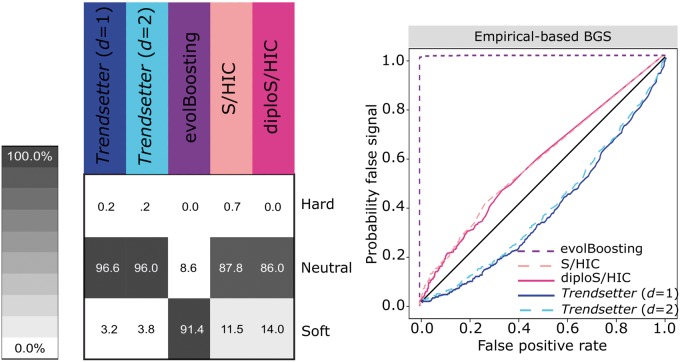
Fig. 7.
Robustness of misclassifying genomic regions undergoing background selection for various methods, under a constant-size demographic history and background selection parameters (number, lengths, and distribution of functional sites as well as distribution of fitness effects) drawn from a distribution based on human data (see Materials and Methods). (Left) Classification rates for regions evolving under background selection. (Right) Probability of misclassifying regions evolving under background selection, by comparing the combined probability of any sweep (hard or soft) under simulations with background selection (probability of false signal) to the same probability under neutral simulations (false positive rate). All methods were trained with three classes: neutral, hard sweep, and soft sweep, with selection coefficients for sweep scenarios drawn uniformly at random on a log scale of .