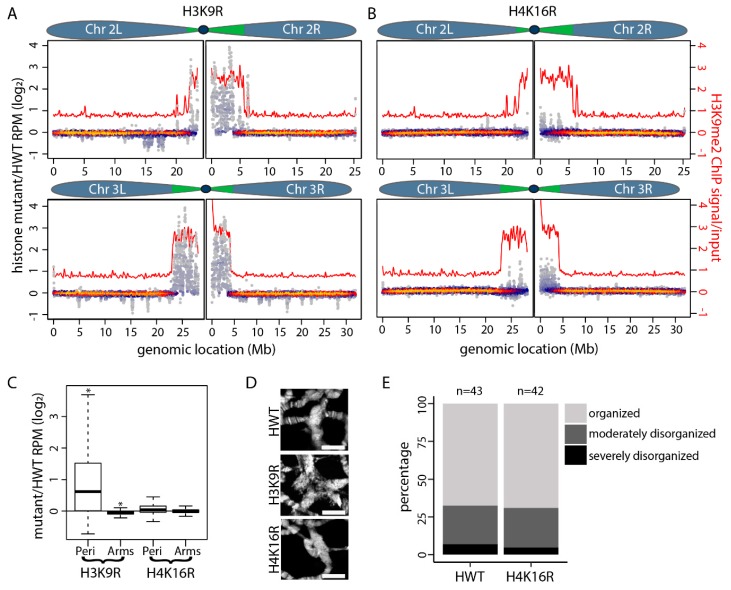
Figure 2.
DNA copy number in pericentric heterochromatin is elevated in H3K9R mutants. (A,B) Heatscatter plot of (A) H3K9R/HWT log2 ratio and (B) H4K16R/HWT log2 ratio of normalized copy number at 10kb windows along Chromosomes 2 and 3. LOESS regression line of modENCODE H3K9me2 ChIP signal is shown in red (GSE47260). (C) Quantification of mutant/HWT ratio of normalized copy number at 10kb windows for all major chromosome scaffolds (Chromosomes 2L, 2R, 3L, 3R, 4 and X) separated into pericentromeres (Peri) and chromosome arms (Arms) (* = p < 0.001; Student’s T test). Coordinates for pericentromeres and chromosome arms were defined in References [52,53] (see also Figure S1). (D) Representative polytene chromosome chromocenter from HWT, H3K9R and H4K16R wandering third-instar salivary glands stained with DAPI. Scale bar = 10 µM. (E) Quantification of cytological categories for HWT and H4K16R chromocenters as performed in Reference [48]. HWT and H4K16R chromocenters shown in panel D represent the organized category whereas the H3K9R chromocenter shown represents the severely disorganized category, which we previously reported comprises 72% of H3K9R chromocenters [48]. The distribution of chromocenters among the three categories between HWT and H4K16R is not statistically different (p > 0.0001; Chi squared test).