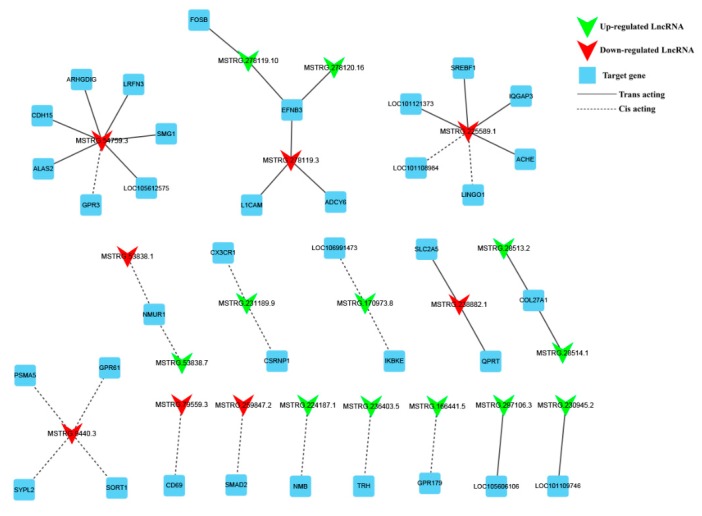
Figure 6.
20 DE lncRNAs and their predictably interactive 36 cis- and trans-targeted genes were composed of this interactive network. The red and green color represent up- and down-regulation, quadrilaterals represent lncRNA, the box represents targeted genes, the straight line and dotted line represent the interaction relationship with trans-and cis-regulation, respectively. CDH15, Cadherin-15; ARHGDIG, Rho GDP dissociation inhibitor γ; LRFN3, leucine rich repeat and fibronectin type III domain containing 3; SMG1, suppressor of morphogenesis in genitalia 1; ALAS2, 5′-aminolevulinate synthase 2; GPR3, G protein-coupled receptor 3; FOSB, FosB proto-oncogene, AP-1 transcription factor subunit; EFNB3, ephrin B3; L1CAM, L1 cell adhesion molecule; ADCY6, adenylate cyclase 6; SREBF1, sterol regulatory element binding transcription factor 1; IQGAP3, IQ motif containing GTPase activating protein 3; ACHE, acetylcholinesterase; LINGO1, leucine rich repeat and Ig domain containing 1; NMUR1, neuromedin U receptor 1; CXCR1, C-X-C motif chemokine receptor 1; CSRNP1, cysteine and serine rich nuclear protein 1; IKBKE, inhibitor of nuclear factor kappa B kinase subunit epsilon; SLC2A5, solute carrier family 2 member 5; QPRT, quinolinate phosphoribosyl transferase; PSMA5, proteasome subunit α 5; GPR61, G protein-coupled receptor 61; SYPL2, synaptophysin like 2; SORT1, sortilin 1; CD69, CD69 molecule; SMAD2, SMAD family member 2; NMB, neuromedin B; TRH, thyrotropin releasing hormone; GPR179, G protein-coupled receptor 179.