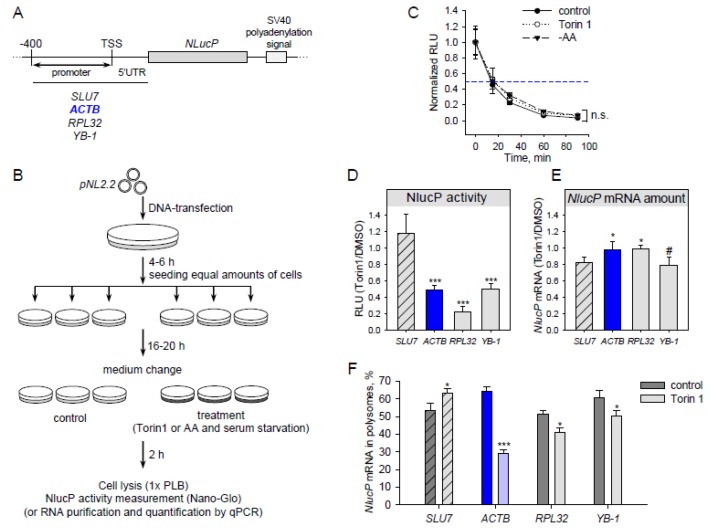
Figure 1.
Translation of reporter mRNA with β-actin 5’UTR is sensitive to mTOR (mammalian target of rapamycin) inhibition. (A) Scheme of the pNL2.2 plasmid used in this study (TSS—transcription start site). (B) Scheme of an experiment with pNL2.2. Cells were transfected with pNL2.2; after 4-6 h equal amounts of cells with the same transfection were seeded to smaller dishes to grow for 16-20 h. Cells were treated for 2h with DMSO (control) or 500 nM Torin1 or subjected to amino acid starvation (EBSS—Earle’s balanced salt solution). Two hours later, luciferase activity or mRNA abundance was measured. (C) HEK293T cells were transfected with pNL2.2-SLU7-NlucP; 24 h later the cells were treated with DMSO (control) or 500 nM Torin1 or EBSS (-AA) for 2 h, then CHX was added for 0, 15, 30, 60, or 90 min. After that, luciferase activity was measured. For each condition, the luciferase activity (Y-axis) was normalized to the value at the zero min time point. (D–F) HEK293T cells were transfected with pNL2.2. After 2h cell treatment with DMSO (control) or 500 nM Torin1 they were collected and luciferase activity (D), NlucP mRNA abundance (E), and polysome distribution (F) were measured. A polysome fraction was calculated as (mRNA in polysome)/(mRNA in polysome + mRNA in subpolysome). Values are the means of at least three independent experiments. Error bars show standard deviations. Student’s t-test was used to estimate the statistical significance versus the SLU7 control (D,E) or versus control treatment (C,F), *** p < 0.001; * p < 0.05; #, n.s., nonsignificant.