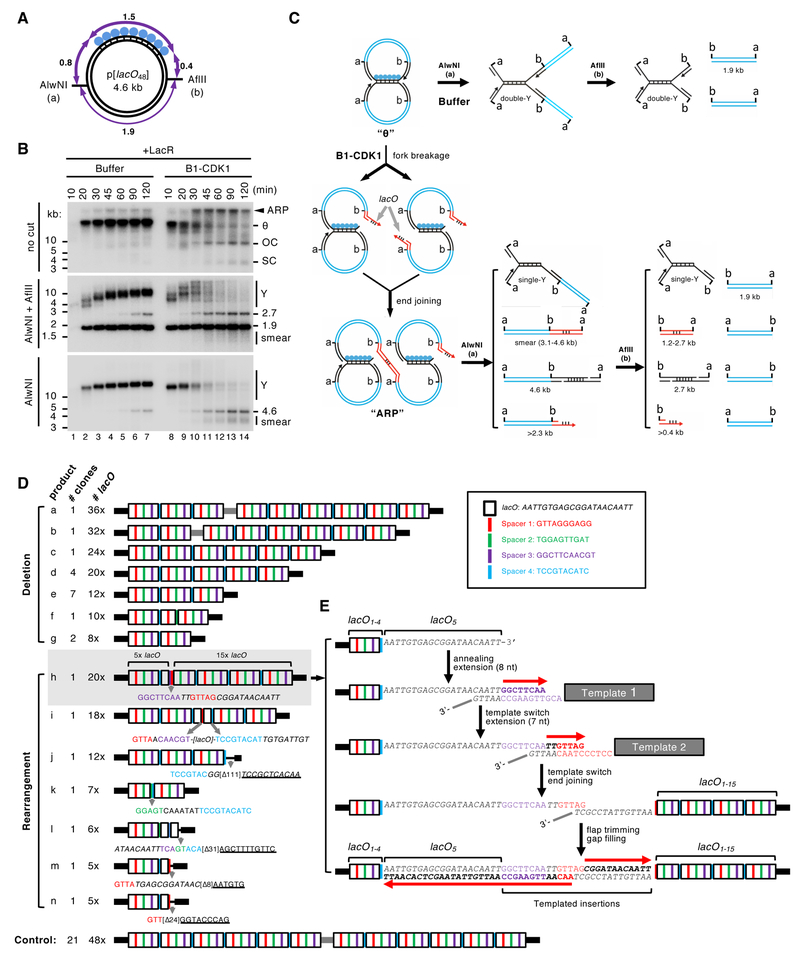
Figure 2. Mitotic processing of stalled replication forks leads to complex DNA rearrangements.
(A) Structure of the 4.6 kb p[lacO48] plasmid. Numbers mark the length of the indicated DNA segments in kilo-basepairs (kb).
(B) p[lacO48] was replicated in the presence of Buffer or B1-CDK1. At the indicated time points, replication products were isolated and digested with AlwNI and AflII, or AlwNI, as indicated. Numbers label the size of linear fragments in kb; Y, double-Y or single-Y structure (see panel C).
(C) Model explaining the restriction products observed in (B). Although the model favors fork breakage on the leading strand, the possibility of fork breakage on the lagging strand has not been excluded. A more detailed model is presented in Figure S2A.
(D) The smear of ~3–4 kb mitotic DNA replication products generated after AlwNI digestion in (B) was self-ligated, cloned and sequenced. The controls are replication products of the same plasmid from a mitotic reaction lacking LacR. The lacO repeats, shown as white boxes, are separated by four unique spacers shown in different colors. Inset, DNA sequences of the lacO repeat and four spacers. The detailed structure of the entire lacO array is shown in Figure S2C.
(E) A model for the generation of product h in (D) from multiple template-switching events.
See also Figure S2.