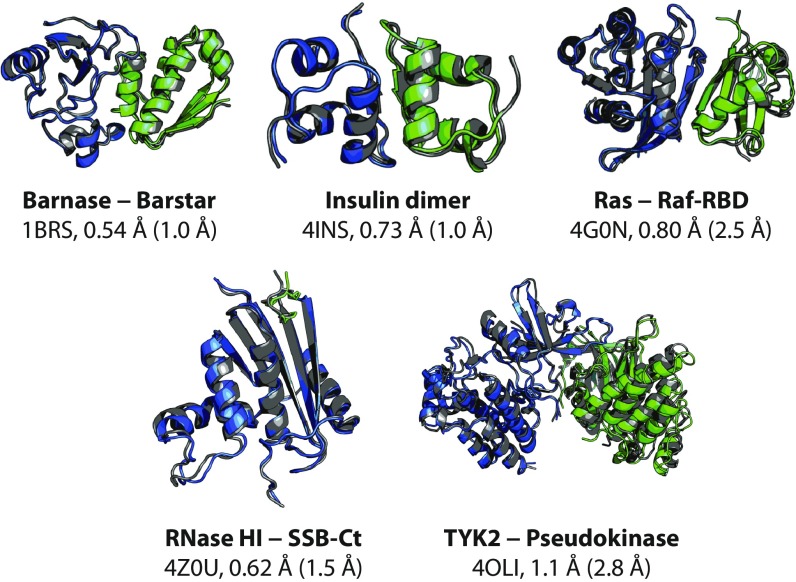
Fig. 1.
The most thermodynamically stable complex visited during reversible-association simulations agrees with the experimentally determined complex within atomic resolution. Representative structures of the most thermodynamically stable complexes observed in reversible-association simulations are shown. For each protein–protein complex, we show a representative associated structure obtained from simulation (blue and green) superimposed on the experimentally determined crystal structure (gray) by a best-fit Cα alignment of the larger protein monomer (blue), along with the name of the complex, the Protein Data Bank (PDB) entry of the experimental structure (3, 26, 50–52), and the Cα interface and ligand RMSDs (I-RMSD and L-RMSD) between the two structures. The protein–protein interface is defined as any pair of Cα atoms, one from each protein monomer, within 10 Å of each other in the experimentally determined complex. The I-RMSD is then calculated by aligning the interface Cα atoms of the representative structure and the experimentally determined structure and determining the Cα RMSD of the interface. The L-RMSD is calculated by first aligning the Cα atoms of the larger protein monomer and then calculating the Cα RMSD of the smaller protein monomer (green) (1). Tempered binding simulations for these five protein–protein systems used the Amber ff99SB*-ILDN (37–39) force field and the TIP3P (40) water model. The representative structure was obtained by clustering the simulations, to avoid bias toward the experimentally determined structure. Clustering was only performed on simulation frames sampled at the lowest tempering rung (rung 0), where the distribution of states is the same as that of a conventional MD simulation. The representative structure from the cluster with the greatest occupancy is used in the figure. Because the tempered binding approach scales all interactions uniformly, the simulations were not biased for, or steered toward, any particular protein–protein complex. Although tempered binding successfully recapitulated experimentally determined bound structures for these systems, its computational expense greatly exceeds that of other approaches, such as docking, for this particular task. We note, however, that its accuracy for this set of five complexes is considerably better than a current state-of-the-art docking program, particularly in its ability to select the correct native-like structure among various low-energy protein–protein complexes (SI Appendix, Table S3), speaking both to the level of sampling achieved by tempered binding and to the accuracy of current MD force fields. Additional descriptions of the systems and the methods are available in SI Appendix.