Fig. 2.
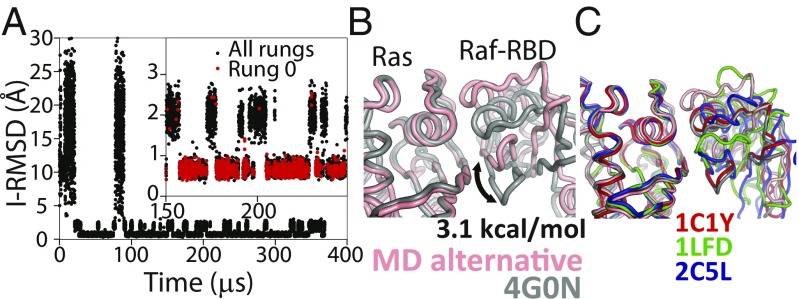
Tempered binding provides a direct atomic-level observation of the ensemble of bound states involved in protein–protein interactions. (A) An I-RMSD trace of a tempered binding simulation of the Ras protein binding to the Ras-binding domain of the Raf effector protein (Raf-RBD) shows reversible association to the crystal complex (PDB ID code 4G0N) (51). For Ras–Raf-RBD, the simulation not only reached the known crystal–structure complex, which was the most thermodynamically stable state, but also an alternative complex about 2 Å away from the crystal structure. The Inset shows a portion of the RMSD trace zoomed in along the y axis. Black (red) circles are points from all (rung 0) trajectory frames. (B) A structure of the alternative state (pink) overlaid onto the crystal structure (gray), aligned to the Ras domain, is shown. Counting the population of the crystal-like state versus the alternative state in rung 0 suggests that the alternative state is ∼3.1 kcal⋅mol−1 higher in free energy than the crystal-like state. (C) An overlay of other Ras–effector complexes demonstrates that the conformation of this alternative state is well within the range of observed binding-interface conformations (53–55).