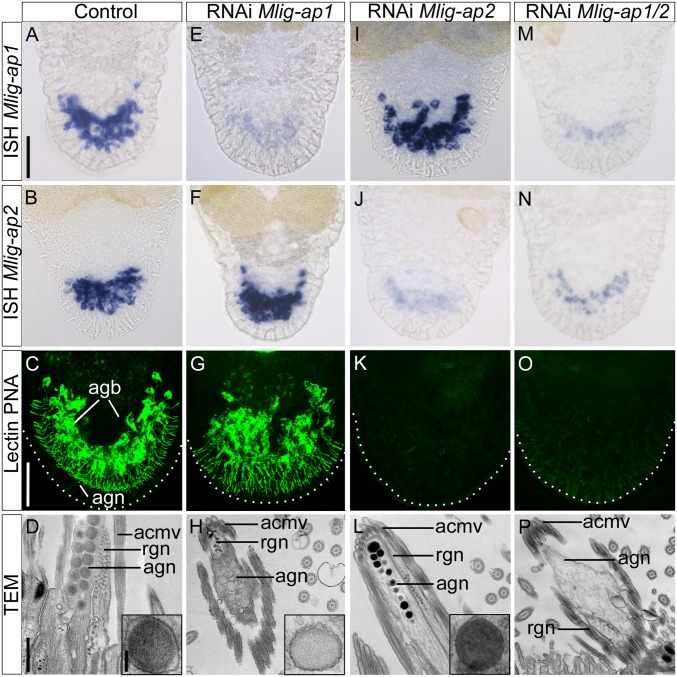
Fig. 3.
RNAi knockdown of adhesive proteins. (A–D) Control animals: in situ hybridization of adhesive proteins (A and B), lectin PNA labeling (confocal projection) (C), and ultrastructural analysis with detail of the adhesive vesicle (TEM) (D). Note the electron-lucent periphery and the electron-dense core of the vesicle (D, Inset). (E–H) Mlig-ap1 RNAi: in situ hybridization of adhesive proteins (E and F), lectin PNA labeling (confocal projection) (G), and ultrastructural analysis with detail of the adhesive vesicle (TEM) (H). Note the lack of the dense core of the vesicle (H, Inset). (I–L) Mlig-ap2 RNAi: in situ hybridization of adhesive proteins (I and J), lectin PNA labeling (confocal projection) (K), and ultrastructural analysis with detail of the adhesive vesicle (TEM) (L). Note the lack of the electron-lucent periphery of the vesicle (L, Inset). (M–P) Double RNAi: in situ hybridization of adhesive proteins (M and N), lectin PNA labeling (confocal projection) (O), and ultrastructural analysis (TEM). Note the complete absence of adhesive vesicles in the adhesive gland cell (P). Dotted lines in confocal images indicate the border of the tail plate. acmv, anchor cell microvilli; agb, adhesive gland body; agn, adhesive gland neck; rgn, releasing gland neck. [Scale bars, 40 µm (A and C), 500 nm (D), and 100 nm (D, Inset).]