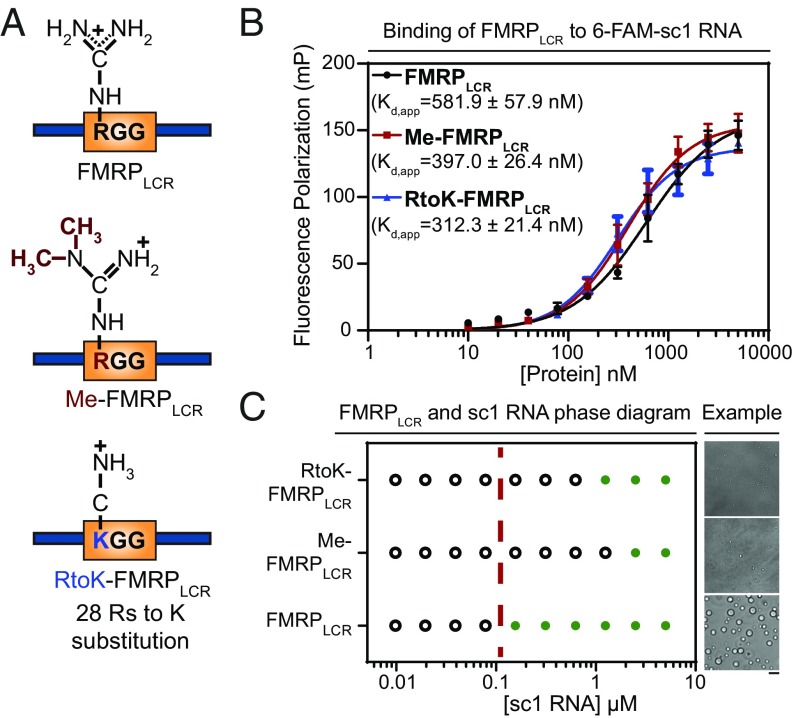
Fig. 5.
Arginine perturbations disrupt FMRPLCR-sc1 RNA phase separation but do not notably affect FMRPLCR binding to sc-1 RNA. (A) Schematic illustration of the three constructs used. (B) Similar binding affinities for interactions between FMRPLCR, Me-FMRPLCR, and RtoK-FMRPLCR with 6-FAM–labeled sc1 RNA (25 nM) are shown using FP assayed in 25 mM Na2PO4, pH 7.4, 100 mM KCl, 2 mM DTT, 0.02 mg/mL E. coli tRNA, and 0.01% Nonidet P-40. Apparent dissociation constants (kd,app ± SD) determined through Hill binding equation (SI Appendix, Eq. S2) from three experimental replicates. (C) Phase diagram of FMRPLCR, Me-FMRPLCR, and RtoK-FMRPLCR at a constant protein concentration of 100 μM with increasing sc1 RNA concentrations in buffer containing 25 mM Na2PO4, pH 7.4, 30 mM KCl, and 2 mM DTT. Green circles represent an observation of droplet formation, and open circles represent no droplet formation detected. The dotted red line is a visual aid for a qualitative phase boundary of FMRPLCR. (Right) Representative DIC microscopy images of droplets for the addition of 2.5 μM of RNA in the conditions described. (Scale bar, 5 μm.) Phase-separation propensity is dramatically decreased for methylated and RtoK mutant proteins, with smaller droplet sizes observed.