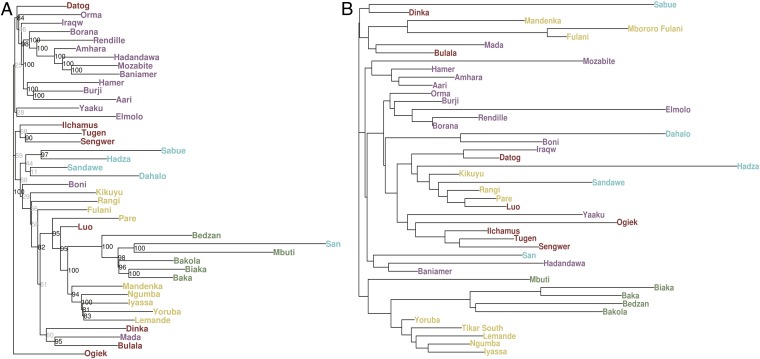
Fig. 2.
Population trees. (A) An NJ population tree was inferred using estimates of pairwise genetic distances between populations based on FST values scaled by Ne. Populations largely cluster by geography or language affiliation, with the notable exceptions of the clade consisting of the Hadza, Sabue, Sandawe, and Dahalo and the clade consisting of the WRHG, ERHG, and San, whose populations cluster together despite being geographically distant. (B) An NJ population tree based on pairwise distances based on the ratio of within-population to between-population haplotype sharing (i.e., IBD); this statistic is more sensitive to recent demographic events, such as gene flow than FST. The EHG cluster most closely with neighboring populations.