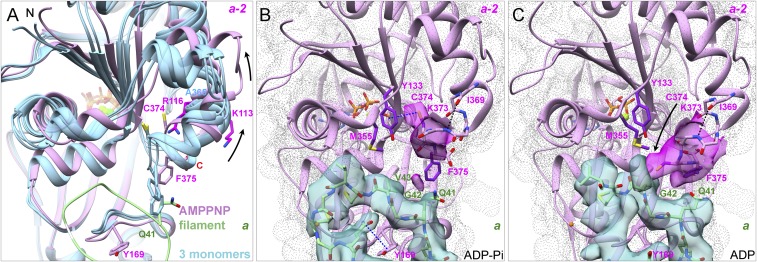
Fig. 6.
Interactions of the C terminus of subunit a-2 with the D-loop of subunit a. (A) Ribbon diagrams comparing the C-terminal regions of our AMPPNP-actin filament structure (plum) with crystal structures (blue) of three actin monomers (Ca-ATP-actin with DNase I, PDB ID code 2A42; Mg-ADP-actin with human gelsolin segment 1, PDB ID code 3A5L; and Mg-ATP-actin with human gelsolin segment 1, PDB ID code 1YAG). F375 clashes with the D-loop of subunit a (green) in two monomers but not in the filament (arrows). (B and C) Experimental densities are semitransparent surfaces in turquoise for subunit a and in magenta for subunit a-2. The gray dot surface for subunit a-2 was calculated from models. Models are rendered as ribbon diagrams and stick figures for the backbone of residues 369–375 and the side chains of C374 and F375. (B) ADP-Pi-actin filament with an intramolecular SH–π interaction between the side chain of C374 and the ring of Y133 of subunit a-2. The side chain of F375 is buried in a hydrophobic pocket formed by P109, L110, I136, V139, A170, P172, and I175 of its subunit and V43 of subunit a. (C) After phosphate release in the ADP-actin filament, the side chain of C374 of subunit a-2 moves ∼8 Å close to the side chain of M355 of its own subunit and forms hydrogen bonds with the backbone of G42 or V43 of subunit a.