Fig. 8.
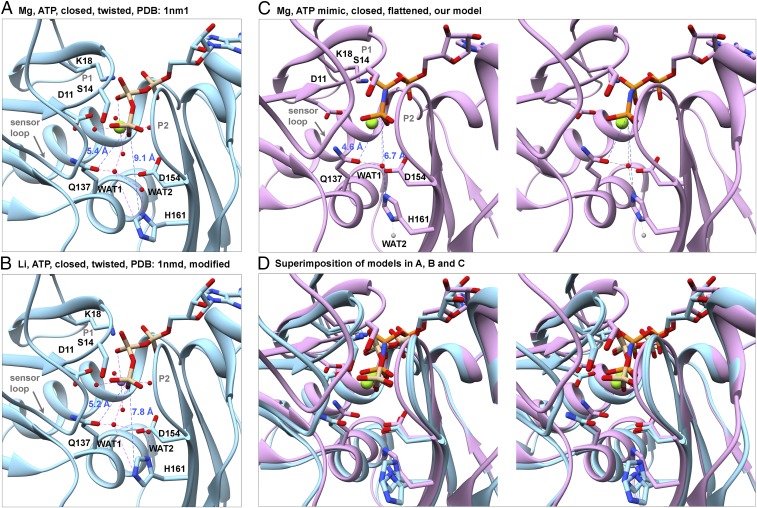
Rearrangement of the catalytic center stimulates ATP hydrolysis by polymerized actin. Ribbon diagrams of models with stick figures of the nucleotides and selected side chains compare distances between the γ-phosphate (Pγ) and the OE1 atom of Q137, NE2 atom of H161, and water molecules in actin monomers and filaments. (A and B) Crystal structure at 1.8 Å resolution of the Dictyostelium Mg-ATP-actin monomer complexed with human gelsolin segment 1 (PDB ID code 1NM1). (B) Based on an analysis of the water network, we flipped the imidazole ring of H161, which fits in the electron density equally well as its conformation in the original PDB file. This change brings the side chain ND1 atom closer to WAT2 (distance: 2.5 Å), indicating that they are hydrogen bonded. (C) Stereoview of our model of polymerized Mg-AMPPNP-actin. Compared with monomers, Pγ is 0.8 Å closer to the OE1 of Q137 and 2.4 Å closer to the NE2 atom of H161, which is rotated by ∼120° in all three filaments relative to monomers (Fig. 7). Based on the direction of protruding density from the imidazole ring of H161 in Fig. 7B, we propose that WAT1 from the X-ray structure (red ball) remains close to OE1 of Q137 and that WAT2 moves away, either downward (gray ball) or upward (not shown). (D) Stereo pair superimposing the models in A–C aligned on subdomain 3 to show their differences.