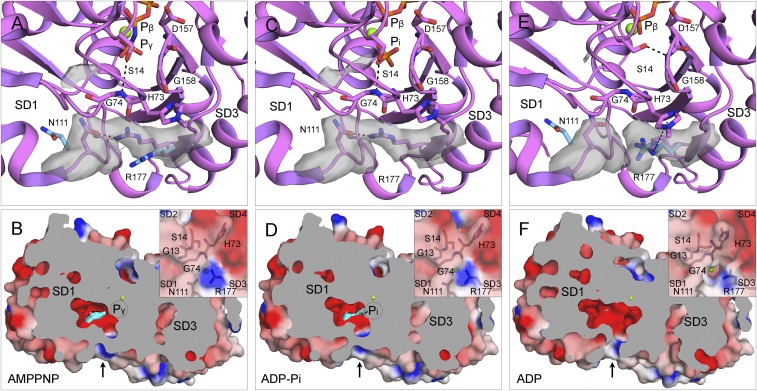
Fig. 9.
Mechanism of phosphate (Pi) release. (A, C, and E) Ribbon diagrams and (B, D, and F) cross-section views with surface representations of single subunits from filaments of AMPPNP-actin (A and B), ADP-Pi-actin (C and D), and ADP-actin (E and F). Ribbon diagrams include stick figures for nucleotides and selected residues, green spheres for Mg, and gray electron potential densities of N111 and R177 and extra density in the Pi cavity. The electrostatic potential surfaces in the cross-section view are rendered with PyMOL using a solvent radius of 1.0 Å with the extra density in the Pi cavity in cyan. (A and C) The S14 side chain forms an H-bond with the backbone of G74, and the side chains of N111 and R177 form an H-bond (∼2.5 Å) that closes the backdoor for Pi release (arrow in B and D). (A and E) The light blue side chains of N111 and R177 are separated by ∼10.0 Å in actin monomers [Ca-ATP-actin, PDB ID code 2A42 (A); Mg-ADP-actin, PDB ID code 3A5L (E)]. (E and F) After phosphate release, the pore of the tunnel lined by the backbone C=O of H73, side chain C=O of N111, and the guanidino group of R177 is open, because the side chain of R177 is turned close enough (<6.0 Å) to the side chain of largely unprotonated methylated H73 to form an N+-H⋅⋅⋅π interaction.