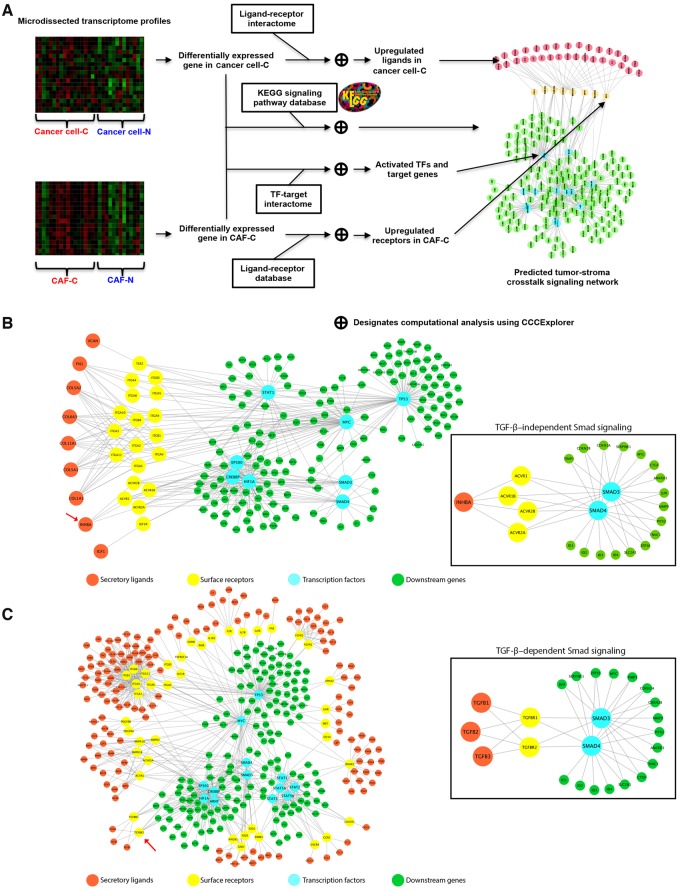
Figure 2.
Identification of stromal–epithelial crosstalk signaling by the CCCExplorer. A) Schematic diagram showing the workflow for the identification of differentially activated signaling pathways in ovarian cancer patients with cancer-associated fibroblast (CAF)–C and CAF-N signatures and the cellular crosstalk with the corresponding cancer cells (cancer cell–C and cancer cell–N, respectively) using transcriptome profiles generated from microdissected ovarian cancer cells and CAFs at the stromal–epithelial interface from the same patient. Such an intercellular crosstalk identification cannot be achieved by using transcriptome profiles generated from bulk tumor tissue alone. To delineate the crosstalk between CAFs and ovarian cancer cells and to identify the signaling pathways contributing to the poor survival rates in the CAF-C patient cohort, new computational functions were added to the multicellular systems biology software CCCExplorer to discover 1) activated transcription factors (TFs) based on increased expression levels of downstream target genes revealed by the transcriptome profile, 2) activated ligand–receptor interaction based on the expression levels of secretory ligands, cell surface receptors, or both, and 3) reconstruction of crosstalk signaling between cell types by linking activated ligand–receptor and TF–downstream target gene information. B) Microdissected transcriptome profiles of CAFs and epithelial tumor cell samples from CAF-C and CAF-N subtype patients were analyzed. Activated signaling pathways in CAF-C were predicted through the identification of overexpressed secretory ligands in cancer cells and activated transcription factors in CAFs in the CAF-C patient cohort. C) Similarly, activated signaling pathways in CAF-C were predicted through the identification of overexpressed receptors and activated transcription factors in CAFs in the CAF-C patient cohort. CAF = cancer-associated fibroblast; TF = transcription factor.