We found the metabolicQTL rs174548 reported by previous metabolites genome-wide association study (GWASs) was significantly associated with lung cancer risk. Based on the concept of mendelian randomization, we proposed that higher levels of serum PUFAs should be causal risk factors of lung cancer.
Abstract
Quantitative trait loci (QTLs) are widely used as instruments to infer causal risk factors of diseases based on the idea of mendelian randomization. Plasma metabolites can serve as risk factors of cancer, and the heritability of many circulating metabolites was high. We conducted a metabolome-wide association study (MWAS) to systematically investigate the effects of genetic variants on metabolites and lung cancer based on published genome-wide association study (GWASs) and metabolic-QTL (mQTL) study. Then we confirmed the results by subsequent genetic and metabolic validations and inferred the causal relationship between identified metabolites and lung cancer through genetic variant(s). We firstly identified six polyunsaturated fatty acids (PUFAs) represented by rs174548-linked haplotype were significantly associated with lung cancer risk in a Chinese GWAS (2311 cases and 3077 controls). Rs174548 was further confirmed to be associated with lung cancer in 13 821 Europeans and 18 471 Asians (ORmeta = 0.87, Pmeta = 1.76 × 10–15) and the effect was much stronger in females (Pinteraction = 6.00 × 10–4). We next validated rs174548-plasma PUFA association in 253 Chinese subjects (β = −0.57, P = 1.68 × 10–3). Rs174548 was also found associated with FADS1 (the major fatty acid desaturase of identified PUFAs) expression in liver tissues. Taken together, we found that rs174548 was associated with both PUFAs and lung cancer. Because rs174548 was the only mQTL variant of PUFAs reported by previous GWASs and explained a large proportion of heritability, we proposed that plasma PUFAs could be causally associated with lung cancer based on the idea of mendelian randomization. These findings provide a diet-related risk factor and may have important implications for prevention on lung cancer.
Introduction
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer-related mortality worldwide. Tobacco consumption is the major cause of lung cancer (1,2); however, the incidence rate of lung cancer is relatively high in Chinese women despite a low prevalence of smoking (3), suggesting the existence of additional predisposing factors. Some lifestyle factors, such as diet, were difficult to quantify accurately in traditional epidemiological studies. Human metabolic phenotypes, which are determined by both genetic and environmental factors, can serve as functional intermediate traits of these factors (4) and help us identify additional risk factors of diseases. Although some studies have successfully associated circulating metabolites with cancer development (5–7), several challenges come up: population-based metabolic studies were limited by specimen availability and cost, with the few published studies of metabolites and diseases being orders of magnitude smaller than studies of diseases alone; commonly using a typical case–control study design, in addition, most studies provided early events or biomarkers but cannot determine a causal relationship between metabolites and diseases. An ordinary prospective study on metabolome may suffer from the biases introduced by metabolic changes because of precancerous lesions or asymptomatic cancers (8), or because of long-term preservation of plasma samples.
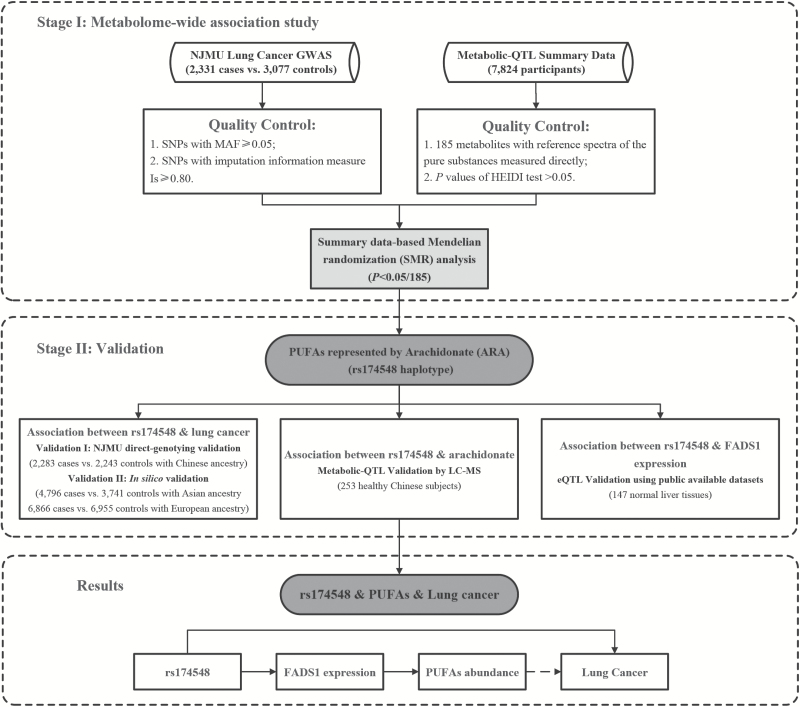
Mendelian randomization (MR) using genetic factors as instrumental variables is widely used for the study to unbiasedly infer causal relationships between exposures and outcomes (9). Two-sample MR is a strategy in which the single nucleotide polymorphism (SNP)-risk factors and SNP-outcome associations are evaluated in different data sources. It helps us avoid the problems mentioned above and allows us to identify metabolic risk factors of diseases from published data of genome-wide association study (GWAS) (10–12). Based on the two-sample MR analysis framework and expression quantitative trait loci (eQTL) information, several transcriptome-wide association studies (TWASs) were successfully applied to indicate functional genes involved in complex diseases (4,13). Similar with eQTL study, a metabolic-QTL (mQTL) study (14) provided a comprehensive atlas of genetic influences on human blood metabolites and reported high contribution of metabolic SNPs to heritability of metabolites recently. Thus, we proposed a metabolome-wide association study (MWAS) to explore the association between metabolites and risk of lung cancer (Figure 1).
Figure 1.
General design of this study.
Materials and methods
Study populations
Based on the published data of circulating metabolite-associated SNPs, we conducted a MWAS to systematically investigate the association between metabolites and lung cancer using our initial NJMU GWAS data (2331 cases and 3077 controls). The association between polyunsaturated fatty acids (PUFAs)-associated rs174548 and lung cancer risk was replicated in 26 884 participants (13 945 cases and 12 939 controls), including direct-genotyping replication (NJMU validation: 2283 cases and 2243 controls) and in silico replication from published GWAS data (MDACC: 1150 cases and 1134 controls; FLCCA: 4796 cases and 3741 controls; NCI: 5716 cases and 5821 controls). Detailed information about the subjects involved in this study is shown in Supplementary Table 1, available at Carcinogenesis Online. NJMU: The NJMU GWAS included 2331 cases and 3077 controls from the Nanjing and Beijing cohorts. The subjects’ details have been reported previously (15). The direct-genotyping replication included 2283 cases and 2243 controls from the Nanjing cohort (749 cases and 754 controls) and the Beijing cohort (1534 cases and 1489 controls). MDACC: A total of 1154 cases of lung cancer and 1137 controls from the MDACC GWAS (16) were obtained from a case–control study at the University of Texas MD Anderson Cancer Center conducted between 1997 and 2007. FLCCA & NCI: We obtained FLCCA and NCI data from the NCBI database of Genotypes and Phenotypes (dbGaP). All samples from the FLCCA (Female Lung Cancer Consortium in Asia) GWAS (17) were female and were non-smokers with Asian ancestry, and the data were downloaded from phs000716.v1.p1. Only data obtained using the Illumina Human660W-Quad v1.0 DNA Analysis BeadChip platform were used for the current analysis, including 4796 cases and 3741 controls. The NCI GWAS included samples from four studies: (i) the Environment and Genetics in Lung Cancer Etiology (EAGLE) study; (ii) the α-Tocopherol, β-Carotene Cancer Prevention Study (ATBC); (iii) the Prostate, Lung, Colon, Ovary Screening Trial (PLCO); and (iv) the Cancer Prevention Study II Nutrition Cohort (CPS-II). We obtained EAGLE study data from dbGaP phs000093.v2.p2, which included 1945 cases and 1992 controls (SLD). We obtained the data of the other three studies from dbGaP phs000336.v1.p1, which included 3782 cases and 3840 controls (CADM). Here, we performed the analyses separately and then conducted a meta-analysis.
An additional cohort of 253 unrelated ethnic Han Chinese was included to evaluate the association between rs174548 and arachidonate (ARA). All subjects were disease-free individuals. Informed consents were obtained from all participants, and the study was approved by the Ethics and Human Subject Committee of Nanjing Medical University with the approval number FWA00001501.
Quality control and imputation of GWAS data sets
The NJMU GWAS was conducted using an Affymetrix Genome-Wide Human SNP Array 6.0 with standard quality-control procedures conducted prior to the association analysis. Though the data sets from the dbGaP were said to be deposited after the initial quality control, we performed standard quality control on the data. We excluded individuals with low call rates (95%), familial relationships and extreme heterozygosity rates, and we excluded SNPs with low call rates (95%), minor allele frequencies (MAFs) <0.05 and P < 1 × 10–6 for the Hardy–Weinberg equilibrium. The data were imputed for the NJMU, FLCCA and NCI GWASs for more than 20 million SNPs using data from the 1000 Genomes Project (the Phase III integrated variant set release, across 2504 samples) as a reference. We phased the haplotypes with Shapeit v2 (http://www.shapeit.fr/, Phasing step) and performed imputations with IMPUTE2 (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html, Imputation step). Poorly imputed SNPs defined by an information measure Is <0.80 with IMPUTE2 were excluded from the analysis.
Metabolome-wide association study for genetic variants using GWAS and mQTL summary data
We applied the Summary data-based Mendelian Randomization (SMR) method (13), which can greatly increase the power for a two-sample MR with large sample sizes, to test for the association between circulating metabolites and lung cancer risk (14). Briefly, SMR method was developed based on the concept of two-sample MR. In this study, let y be a phenotype (lung cancer), x be exposure (abundance of a metabolite), z be an instrumental variable (SNP), we estimated:
(1) the effect of metabolites on lung cancer () through:
Where was the estimate of effect of a SNP on lung cancer from our NJMU lung cancer GWAS and was the estimate of a SNP effect on the abundance of a metabolite from mQTL GWAS.
(2) the sample variance of . var( was estimated by the Delta method (18);
(3) approximate χ2 test statistic to test the significance of : .
Metabolic-QTL summary data was extracted from the association results of a GWAS on the human metabolome conducted by So-Youn Shin et al. (14). The metabolites were relatively quantified (14) by liquid-phase chromatography coupled with tandem mass spectrometry (LC-MS). We obtained the association data from http://metabolomics.helmholtz-muenchen.de/gwas/index.php?task=download, which contains all association statistics for 453 metabolites with a P ≤ 1 × 10–5. We included SNPs associated with 238 metabolites with reference spectra of the pure substances measured directly at the GWAS significance level of P ≤ 5 × 10–8. To minimize the heterogeneity between two GWASs, we further excluded SNPs with MAF<0.05 estimated in both two GWAS data and remained 13 266 SNPs. We performed heterogeneity in dependent instruments (HEIDI) test to distinguish causality from linkage, and excluded SNPs in linkage disequilibrium with the lead metabolic trait-associated SNP at a threshold of r2 > 0.9. The EUR population of the 1000 Genome project was set as the reference to estimate the linkage disequilibrium correlation between SNPs.
Replication genotyping
Genotyping of rs174548 was performed using TaqMan assays (Applied Biosystems). The primers and probes are available upon request. Call rates for SNP genotypes were >95% in each of the replication series. The laboratory technicians who performed the genotyping experiments were blinded to case or control status. Five percent of the samples were randomly selected for repeat genotyping as blind duplicates, and the reproducibility was >99%.
The measurements of plasma ARA
Plasma ARA of 253 subjects was measured using ultra high-performance liquid chromatography tandem with mass spectrometry (LC-MS) according to the previous report (19). Briefly, plasma samples were thawed at 4°C prior to analysis. Then, 100 μl plasma was mixed with 300 μl methanol for protein precipitation, and the supernatant was obtained after centrifugation at 15000×g for 15 min. After drying, the residue was reconstituted in 200 µl water. A total of 10 μl of the supernatant was injected into the Ultimate 3000 system (Dionex) coupled to a Q-Exactive mass spectrometer (Thermo Fisher Scientific) for metabolomic analysis. The ZORBAX SB-C18 column (2.1 × 100 mm) was used, and column temperature was maintained at 40°C. Mobile phase conditions consisted of acetonitrile containing 0.1% formic acid as mobile phase A and water containing 0.1% formic acid as mobile phase B. The gradient program was 0–3 min at 1% mobile phase A, 3–10 min at 1–99% mobile phase A, 10–13 min at 99% mobile phase A and 13–15 min at 1% mobile phase A. The mass spectrometry was run in electrospray positive ion and negative ion full scan mode (110.0–800.0 m/z). Capillary voltage for positive ion mode was −2500 V, and the negative ion mode was 3500 V. The remaining parameters were: temperature 425°C and capillary temperature 300°C. All samples were arranged in random order to avoid biases related to the injection order. In the current study, 25 pooled plasma samples (253 samples) were processed as QC samples and randomly placed in the sample queue to monitor the stability of the LC−MS system. LC-MS raw data were processed using TraceFinder 3.1 (Thermo Fisher Scientific). The metabolite identification was based on the comparison of accurate mass and retention time with 260 standard compounds. As ARA was the PUFA which was most affected by rs174548, we selected ARA to represent the family of PUFAs in the validation procedure.
The expression of FADS1 and FADS2
Expression levels of normal liver tissues were obtained from TCGA Firehose at the MIT Broad Institute (https://confluence.broadinstitute.org/display/GDAC/Home, 2014-07-15 release). Normalized read counts were used to quantify the expression of FADS1 and FADS2. The results and the boxplots of the eQTL analysis from the GTEx project were obtained from GTEx Analysis V6 (dbGaP accession number phs000424.v6.p1). The expression of FADS1 was extracted from the fully processed, normalized and filtered gene expression data released by the GTEx portal.
Selection of EPA-related SNPs and construction of weighted genetic risk score
We included all eicosapentaenoate (EPA)-related SNPs released by a previous study (14) (PEPA < 5 × 10–5) and excluded ARA-SNPs (PARA < 5 × 10–5) in this analysis. When multiple SNPs were observed in strong linkage disequilibrium (r2 ≥ 0.8), only the SNP with the lowest metabolites–SNP association P value was selected. A total of 11 SNPs were included in the analysis and the associations between the 11 SNPs and lung cancer were listed in Supplementary Table 2, available at Carcinogenesis Online. We then derived a weighted genetic risk score (wGRS) predicting serum EPA level by summing the dosages for EPA-increasing alleles with 11 EPA-specific SNPs (PEPA < 5 × 10–5 and PARA ≥ 5 × 10–5) using the following formula:, where is the effect estimate of the th SNP for serum EPA level released by a previous study (14) and SNP is the dosage of the effect allele (0, 1 or 2). Logistic regression model was used to estimate odds ratios (OR) and 95% confidence intervals for associations between genetically predicted plasma EPA level and lung cancer risk.
Other statistical analyses
For the genetic studies, we tested SNPs for associations with lung cancer using logistic regression analysis in PLINK 1.07 (20). Tests of association between imputed SNPs and lung cancer were performed under a probabilistic dosage model in SNPTEST v2.5 (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/ snptest.html). The meta-analysis was performed using inverse-variance approaches. ORs and 95% confidence intervals were calculated by logistic regression analysis under an additive model, with adjustments for age, gender, smoking status and corresponding principal components. For the CADM study from the NCI project, we did not include the smoking status in the model since such information was not provided. In the stratification analysis, we calculated Cochran’s Q statistic to test for heterogeneity between the groups. A linear regression model was used to perform QTL analysis. The plasma ARA values and the expression levels of genes used in the model were log2 transformed. The R package ggplot2 (21) was used to create the association plots. General analyses were performed with R software (version 3.1.1).
Results
Metabolome-wide association study
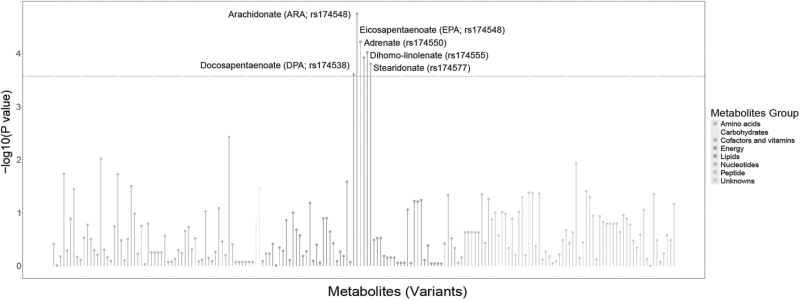
We applied a SMR method to test the association between circulating metabolites and the risk of lung cancer. We included 13 266 metabolites-associated SNPs in the initial scan, representing 238 metabolites. A total of 185 SNP-metabolite pairs passed the test for HEIDI (Phet > 0.05) and were included in further analysis (Figure 2). After correction for multiple testing (P < 0.05/185), arachidonate (ARA; 20:4n6) (P = 1.79 × 10–5), eicosapentaenoate (EPA; 20:5n3) (P = 6.03 × 10–5), dihomo-linolenate (20:3n3 or n6) (P = 9.58 × 10–5), adrenate (22:4n6) (P = 1.20 × 10–4), stearidonate (18:4n3) (P = 1.57 × 10–4) and docosapentaenoate (n3 DPA; 22:5n3) (P = 2.63 × 10–4) were found to be associated with a significantly increased risk of lung cancer (Supplementary Table 3, available at Carcinogenesis Online, Figure 2). All the metabolites above were PUFAs, and all the variants associated with these PUFAs were located in a single haplotype (Supplementary Figure 1, available at Carcinogenesis Online). Because the effect of rs174548 on lung cancer risk was the strongest, we used it as the instrumental variant for six PUFAs in following analysis.
Figure 2.
Results of SMR tests for the association between metabolites and lung cancer in NJMU GWAS. The presented summary statistics are form the summary-data based mendelian randomization (SMR) tests conducted in 2331 cases and 3077 controls of Chinese ancestry. Each point represents a SNP-metabolite pair with its log10-transformed P value. The colors coding of all metabolites indicates eight broad metabolic groups. The dash green line indicates the statistically significant P value threshold after Bonferroni correction for multiple-hypothesis testing (P < 0.05/185).
rs174548 was consistently associated with lung cancer and had stronger effect in females
We validated the association between rs174548 and lung cancer risk by directly genotyping an additional 2283 cases and 2243 controls (Validation I) in Chinese individuals and by in silico validation based on three lung cancer GWASs including a total of 11 662 cases and 10 696 controls (Validation II) (Table 1, Supplementary Figure 2, available at Carcinogenesis Online). The pooled OR of lung cancer was 0.87 for the G allele of rs174548 (Pcombined = 1.76 × 10–15). Stratification by gender in our lung cancer GWAS and Validation I study suggested that the effect of rs174548 was stronger in females (PQ = 0.01, Supplementary Table 4, available at Carcinogenesis Online). However, no heterogeneity was found between smokers and non-smokers (PQ > 0.05, Supplementary Table 4, available at Carcinogenesis Online). In addition, we investigated the effect of epistasis between rs174548 and gender and identified a significant interaction between the G allele of rs174548 and female gender on lung cancer risk (interaction OR = 0.85, P = 6.00 × 10–4, Supplementary Table 5, available at Carcinogenesis Online).
Table 1.
Summary of discovery and validation studies for rs174548
| Phase | Study | EAa | OAb | EAFc | Allele frequencyd | OR (95% CI) | P e | ||
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | ||||||
| Discovery | NJMU(Nanjing) | G | C | 0.37 | 0.41 | 192.0/702.0/579.0 | 331.0/943.0/688.0 | 0.89(0.79–0.99) | 3.34E−02 |
| NJMU(Beijing) | G | C | 0.34 | 0.41 | 109.0/369.0/380.0 | 182.0/545.0/388.0 | 0.71(0.62–0.82) | 1.46E−06 | |
| Validation I | NJMU Validation (Nanjing) | G | C | 0.35 | 0.40 | 74.0/370.0/293.0 | 124.0/352.0/273.0 | 0.81(0.69–0.96) | 1.22E−02 |
| NJMU Validation (Beijing) | G | C | 0.30 | 0.38 | 150.0/630.0/734.0 | 234.0/669.0/571.0 | 0.68(0.60–0.76) | 1.25E−11 | |
| Validation II | MDACC | G | C | 0.31 | 0.33 | 99.9/510.0/540.1 | 124.1/507.8/502.1 | 0.89(0.77–1.00) | 4.67E−02 |
| FLCCA | G | C | 0.40 | 0.43 | 858.3/2133.7/1804.0 | 731.1/1738.3/1271.6 | 0.91(0.85–0.97) | 4.90E−03 | |
| NCI | G | C | 0.30 | 0.31 | 516.0/2424.3/2775.7 | 582.9/2433.4/2804.7 | 0.93(0.87–0.98) | 1.18E−02 | |
| META | G | C | 0.87(0.84–0.90) | 1.76E−`15 | |||||
aEffect allele.
bOther allele.
cEffect allele frequency.
dEA homozygote/heterozygote/OA homozygote.
eAdjusted by age, gender, smoking status (or py) and PC(s).
rs174548 was a metabolic variant of PUFAs in individuals with Chinese ancestry
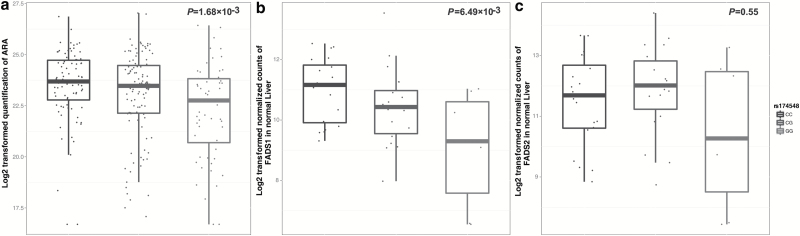
The SNP rs174548 was previously reported to be associated with PUFAs (Supplementary Table 6, available at Carcinogenesis Online) in populations of European ancestry. Thus, we additionally studied the association between rs174548 and PUFAs in 253 healthy Chinese individuals to avoid population heterogeneity. Because abundances of plasma PUFAs were highly correlated and ARA was the PUFA most affected by rs174548 in previous study (Supplementary Table 6, available at Carcinogenesis Online), we selected ARA as an indicator of six identified PUFAs. As expected, carriers of the rs174548 G allele had significantly lower plasma levels of ARA (β = −0.57, P = 1.68 × 10–3) (Figure 3a). These results suggested that the SNP was a mQTL variant in populations of both European and Chinese ancestries.
Figure 3.
Quantitative trait loci analysis of rs174548. (a) The G allele of rs174548 was significantly associated with decreased plasma ARA in 253 individuals with Chinese ancestry (β = −0.57, P = 1.68 × 10–3). (b) The G allele of rs174548 was significantly associated with decreased expression of FADS1 in 50 normal liver tissues from the TCGA project (β = −0.84, P = 6.49 × 10–3). (c) The G allele of rs174548 was not associated with the expression of FADS2 in 50 normal liver tissues from the TCGA project (β = −0.23, P = 0.55). Plasma ARA levels and FADS1/FADS2 expression levels were log2 transformed. The G allele of rs174548 was the minor allele in both Asian and European populations. P value was calculated with the linear regression model. The boxplot displays the first and third quartiles (top and bottom of the boxes), the median (band inside the boxes), and the lowest and highest point within 1.5 times the interquartile range of the lower and higher quartiles (whiskers).
We also performed eQTL analysis on 50 normal liver tissues to study the association between rs174548 and two major desaturases (FADS1 and FADS2), as liver is the major organ that influences circulating PUFA levels (22). The G allele of rs174548 was significantly associated with the decreased expression of FADS1 (β = −0.84, P = 6.49 × 10–3) but not the expression of FADS2 (β = −0.23, P = 0.55) (Figure 3b and c). These results were further validated in 97 normal liver tissues from the GTEx project (β = −0.38, P = 2.20 × 10–3 for FADS1, Supplementary Figure 3, available at Carcinogenesis Online). Then, we further performed a sex-specific eQTL analysis in normal liver tissues from the GTEx project, and we observed a significantly stronger effect in female normal liver tissues than males (P for heterogeneity test = 3.85 × 10–2) (Supplementary Figure 3, available at Carcinogenesis Online).
Discussion
PUFAs are classified into omega-6s and omega-3s (23). Linoleic acid and ɑ-linolenic acid, the precursor of both the omega-6 and omega-3 synthesis pathways (Supplementary Figure 4, available at Carcinogenesis Online), are essential fatty acids, thus both can be obtained through diet. Vegetable oils are major sources of omega-6 fatty acids, whereas seafood is a major source of omega-3 fatty acids (24). Although PUFAs intake was traditionally considered as a protective factor of cardiovascular diseases (25–27), a recent double blind randomized controlled study suggested that replacement of saturated fat with linoletic acid remarkably increase the risk of death (28). For cancer, multiple epidemiological studies have investigated the association between dietary intake of PUFA and the development of cancer (including breast, colorectal, prostate and pancreatic cancer) in prospective studies but failed to achieve consistent results (29). In this study, we first provided evidence for the causal association between in vivo PUFAs and lung cancer risk using a MWAS and subsequent genetic and metabolic validations. Because PUFAs are diet-related, control of PUFA intake may be an easily available and economical way to reduce lung cancer incidence, especially in women.
In this study, rs174548 is an ideal instrumental variant to infer the relationship between PUFAs and lung cancer. According to the previous report (14), estimated heritability of ARA was about 0.30 and rs174548 itself explained approximately 20% of the total heritability. In addition, rs174548 related haplotype is the only one supported by multiple metabolic GWASs (14,30). In liver tissues, our eQTL analysis additionally validated the association between rs174548 and expression of FADS1, which encodes one of the major desaturases of PUFAs identified above (Supplementary Figure 4, available at Carcinogenesis Online). Thus, our results provided a reasonable explanation for association between rs174548 and lung cancer (Figure 1).
However, it is still challenging to determine the functional PUFA underlying the association between rs174548 and lung cancer, because FADS1 encodes a fatty acid desaturase shared by both the omega-6 and omega-3 PUFA synthesis pathways (Supplementary Figure 4, available at Carcinogenesis Online). All circulating PUFAs related to this desaturase are highly correlated (14) and are associated with rs174548 (14). Previous studies have reported that omega-6 and omega-3 PUFAs have opposing effects (24). ARA is the predominant omega-6 fatty acid. ARA can be converted to eicosanoids, which are important regulators of inflammation. Tumor-prompting inflammation is a classic hallmark of cancer (31). The primary metabolic enzyme of ARA, cyclooxygenase (COX), is the target of non-steroidal anti-inflammatory drugs (NSAIDs), which have been reported to have beneficial effects on reducing the risk of developing some solid tumors, including the four most prevalent cancers worldwide: breast, colon, lung and prostate cancer (32). Recent lung cancer GWAS studies in female never-smokers and EGFR mutated patients also identified a series of susceptibility loci in HLA region, providing additional evidence for tumor promoting role in the development of lung cancer. Eicosapentaenoic acid and docosahexaenoic acid (DHA), however, are the typical omega-3 PUFAs, and they can be converted to anti-inflammatory products (24). Recently, experimental studies have provided evidence that the bioactive lipids that come from ARA (eicosanoids) play critical roles in cancer progression (33), while EPA has been frequently reported to have potential effect in cancer prevention (34–39). Thus, we further evaluated the association between EPA and lung cancer with EPA-specific SNPs, and observed that genetically determined plasma EPA level was not significantly associated with lung cancer risk (OR = 0.49, P = 1.29 × 10–1) (Supplementary Table 2, available at Carcinogenesis Online). Thus, omega-6 PUFAs are more likely to be the causal factor of lung cancer rather than omega-3 PUFAs. However, we could not rule out the potential effect of omega-3 PUFAs in the etiology of lung cancer. Additionally, the metabolites were relatively quantified (14) by liquid-phase chromatography coupled with tandem mass spectrometry (LC-MS) in this study, which could not represent the absolute abundance. Hence, further studies are warranted to clarify the causal PUFA(s) and illuminate the underlying mechanisms.
Further analysis stratified by gender revealed that the protective effect of rs174548 was more apparent in females, and the sex-specific eQTL analysis in normal liver tissues also revealed that the association between rs174548 and FADS1 expression was significantly stronger in female normal liver tissues than males (Supplementary Figure 3, available at Carcinogenesis Online). Previous studies have demonstrated that sex hormones may participate in the metabolism of ARA (40). 17β-estradiol, the primary female sex hormone, was reported to facilitate the lipid peroxidation of omega-6 PUFAs by synergistically interacting with omega-6 PUFA metabolism. The lipid peroxidation-derived byproducts would further generate miscoding etheno-DNA adducts, which were potentially harmful carcinogens (41).
In addition, the effect of rs174548 on lung cancer risk was more apparent in Chinese studies (Supplementary Figure 2, available at Carcinogenesis Online). Interestingly, two rs174548-linked common variants (rs174550 and rs174549) have been reported to be associated with the risk of colorectal cancer and laryngeal squamous cell carcinoma in previous Asian GWASs (42,43), whereas no association between rs174548-linked SNPs and cancers have ever been identified in GWASs with individuals of European ancestry. Because diet is the major source of omega-6 PUFAs, varied dietary habits may be one of the reasons for this difference. A systematic nutrition survey including 1 630 069 individuals from 266 countries indicated that mean dietary omega-6 PUFA consumption is higher in Chinese individuals than in individuals from other countries (44).
Taken together, our study identified mQTL rs174548 at 11q12.2 as a novel and smoking-independent genetic variant associated with lung cancer, particularly in females. With the idea of MR, we proposed that higher levels of plasma PUFAs could be causal risk factors for lung cancer. Because PUFAs are diet-related, these findings may have important implications for prevention on lung cancer.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Funding
This work was supported by National Natural Science of China (81521004, 81230067, 81573238, 81225020, 81703295), National Program for Support of Top-notch Young Professionals from the Organization Department of the CPC Central Committee, Jiangsu Specially-Appointed Professor project, Science Foundation for Distinguished Young Scholars in Jiangsu (BK20160046), the Innovation of Graduate Student Training Project in Jiangsu Province (KYZZ15_0268), the Priority Academic Program for the Development of Jiangsu Higher Education Institutions [Public Health and Preventive Medicine] and Top-notch Academic Programs Project of Jiangsu Higher Education Institutions (PPZY2015A067).
Conflict of Interest Statement: None declared.
Supplementary Material
Abbreviations
- ARA
arachidonate
- eQTL
expression quantitative trait loci
- EPA
eicosapentaenoate
- GWAS
genome-wide association study
- MAF
minor allele frequency
- MWAS
metabolome-wide association study
- mQTL
metabolic-QTL
- MR
mendelian randomization
- OR
odds ratios
- PUFA
polyunsaturated fatty acid
- QTL
quantitative trait loci
- SNP
single nucleotide polymorphism
- SMR
summary data-based mendelian randomization.
References
- 1. Godtfredsen N.S., et al. (2005) Effect of smoking reduction on lung cancer risk. JAMA, 294, 1505–1510. [DOI] [PubMed] [Google Scholar]
- 2. Peto R., et al. (2000) Smoking, smoking cessation, and lung cancer in the UK since 1950: combination of national statistics with two case-control studies. BMJ, 321, 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Torre L.A., et al. (2015) Global cancer statistics, 2012. CA. Cancer J. Clin., 65, 87–108. [DOI] [PubMed] [Google Scholar]
- 4. Gusev A., et al. (2016) Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet., 48, 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mayers J.R., et al. (2014) Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat. Med., 20, 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arslan A.A., et al. (2014) Circulating estrogen metabolites and risk of breast cancer in postmenopausal women. Cancer Epidemiol. Biomarkers Prev., 23, 1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bai L., et al. (2016) An integrated genome-wide systems genetics screen for breast cancer metastasis susceptibility genes. PLoS Genet., 12, e1005989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dang C.V. (2012) Links between metabolism and cancer. Genes Dev., 26, 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith G.D., et al. (2003) ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol., 32, 1–22. [DOI] [PubMed] [Google Scholar]
- 10. Burgess S., et al. ; EPIC- InterAct Consortium. (2015) Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol., 30, 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evans D.M., et al. (2015) Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu. Rev. Genomics Hum. Genet., 16, 327–350. [DOI] [PubMed] [Google Scholar]
- 12. Lopez-Maestre H., et al. (2016) SNP calling from RNA-seq data without a reference genome: identification, quantification, differential analysis and impact on the protein sequence. Nucleic Acids Res., 44, e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu Z., et al. (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet., 48, 481–487. [DOI] [PubMed] [Google Scholar]
- 14. Shin S.Y., et al. ; Multiple Tissue Human Expression Resource (MuTHER) Consortium. (2014) An atlas of genetic influences on human blood metabolites. Nat. Genet., 46, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu Z., et al. (2011) A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat. Genet., 43, 792–796. [DOI] [PubMed] [Google Scholar]
- 16. Amos C.I., et al. (2008) Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet., 40, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lan Q., et al. (2012) Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat. Genet., 44, 1330–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lynch M., et al. (1998) Genetics and Analysis of Quantitative Traits. Sinauer, Sunderland, MA. [Google Scholar]
- 19. Chen M., et al. (2013) Association of exposure to phenols and idiopathic male infertility. J. Hazard. Mater., 250-251, 115–121. [DOI] [PubMed] [Google Scholar]
- 20. Purcell S., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ginestet C. (2011) ggplot2: Elegant Graphics for Data Analysis. J. Roy. Stat. Soc. Series A Stat. Soc., 174, 245–245. [Google Scholar]
- 22. Rapoport S.I., et al. (2010) Quantitative contributions of diet and liver synthesis to docosahexaenoic acid homeostasis. Prostaglandins. Leukot. Essent. Fatty Acids, 82, 273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chilton F.H., et al. (2014) Diet-gene interactions and PUFA metabolism: a potential contributor to health disparities and human diseases. Nutrients, 6, 1993–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmitz G., et al. (2008) The opposing effects of n-3 and n-6 fatty acids. Prog. Lipid Res., 47, 147–155. [DOI] [PubMed] [Google Scholar]
- 25. Harris W.S., et al. (2009) Omega-6 fatty acids and risk for cardiovascular disease: a science advisory from the American Heart Association Nutrition Subcommittee of the Council on Nutrition, Physical Activity, and Metabolism; Council on Cardiovascular Nursing; and Council on Epidemiology and Prevention. Circulation, 119, 902–907. [DOI] [PubMed] [Google Scholar]
- 26. Kromhout D., et al. (1985) The inverse relation between fish consumption and 20-year mortality from coronary heart disease. N. Engl. J. Med., 312, 1205–1209. [DOI] [PubMed] [Google Scholar]
- 27. Lavie C.J., et al. (2009) Omega-3 polyunsaturated fatty acids and cardiovascular diseases. J. Am. Coll. Cardiol., 54, 585–594. [DOI] [PubMed] [Google Scholar]
- 28. Ramsden C.E., et al. (2016) Re-evaluation of the traditional diet-heart hypothesis: analysis of recovered data from Minnesota Coronary Experiment (1968-73). BMJ, 353, i1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Azrad M., et al. (2013) Current evidence linking polyunsaturated fatty acids with cancer risk and progression. Front. Oncol., 3, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu Y., et al. (2016) Genome-wide meta-analyses identify novel loci associated with n-3 and n-6 polyunsaturated fatty acid levels in Chinese and European-ancestry populations. Hum. Mol. Genet., 25, 1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hanahan D., et al. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 32. Harris R.E. (2009) Cyclooxygenase-2 (cox-2) blockade in the chemoprevention of cancers of the colon, breast, prostate, and lung. Inflammopharmacology, 17, 55–67. [DOI] [PubMed] [Google Scholar]
- 33. Wang D., et al. (2010) Eicosanoids and cancer. Nat. Rev. Cancer, 10, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bartsch H., et al. (1999) Dietary polyunsaturated fatty acids and cancers of the breast and colorectum: emerging evidence for their role as risk modifiers. Carcinogenesis, 20, 2209–2218. [DOI] [PubMed] [Google Scholar]
- 35. Hardman W.E. (2004) (n-3) fatty acids and cancer therapy. J. Nutr., 134(12 Suppl), 3427S–3430S. [DOI] [PubMed] [Google Scholar]
- 36. Isbilen B., et al. (2006) Docosahexaenoic acid (omega-3) blocks voltage-gated sodium channel activity and migration of MDA-MB-231 human breast cancer cells. Int. J. Biochem. Cell Biol., 38, 2173–2182. [DOI] [PubMed] [Google Scholar]
- 37. Wu M., et al. (2005) Omega-3 polyunsaturated fatty acids attenuate breast cancer growth through activation of a neutral sphingomyelinase-mediated pathway. Int. J. Cancer, 117, 340–348. [DOI] [PubMed] [Google Scholar]
- 38. Schley P.D., et al. (2005) Mechanisms of omega-3 fatty acid-induced growth inhibition in MDA-MB-231 human breast cancer cells. Breast Cancer Res. Treat., 92, 187–195. [DOI] [PubMed] [Google Scholar]
- 39. Connolly J.M., et al. (1999) Effects of reduced dietary linoleic acid intake, alone or combined with an algal source of docosahexaenoic acid, on MDA-MB-231 breast cancer cell growth and apoptosis in nude mice. Nutr. Cancer, 35, 44–49. [DOI] [PubMed] [Google Scholar]
- 40. Decsi T., et al. (2011) Sex-specific differences in essential fatty acid metabolism. Am. J. Clin. Nutr., 94(6 Suppl), 1914S–1919S. [DOI] [PubMed] [Google Scholar]
- 41. Sun X., et al. (2012) Lipid peroxidation and DNA adduct formation in lymphocytes of premenopausal women: Role of estrogen metabolites and fatty acid intake. Int. J. Cancer, 131, 1983–1990. [DOI] [PubMed] [Google Scholar]
- 42. Zhang B., et al. ; Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO); Colorectal Transdisciplinary (CORECT) Study; Colon Cancer Family Registry (CCFR). (2014) Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat. Genet., 46, 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wei Q., et al. (2014) Genome-wide association study identifies three susceptibility loci for laryngeal squamous cell carcinoma in the Chinese population. Nat. Genet., 46, 1110–1114. [DOI] [PubMed] [Google Scholar]
- 44. Micha R., et al. ; Global Burden of Diseases Nutrition and Chronic Diseases Expert Group NutriCoDE. (2014) Global, regional, and national consumption levels of dietary fats and oils in 1990 and 2010: a systematic analysis including 266 country-specific nutrition surveys. BMJ, 348, g2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.