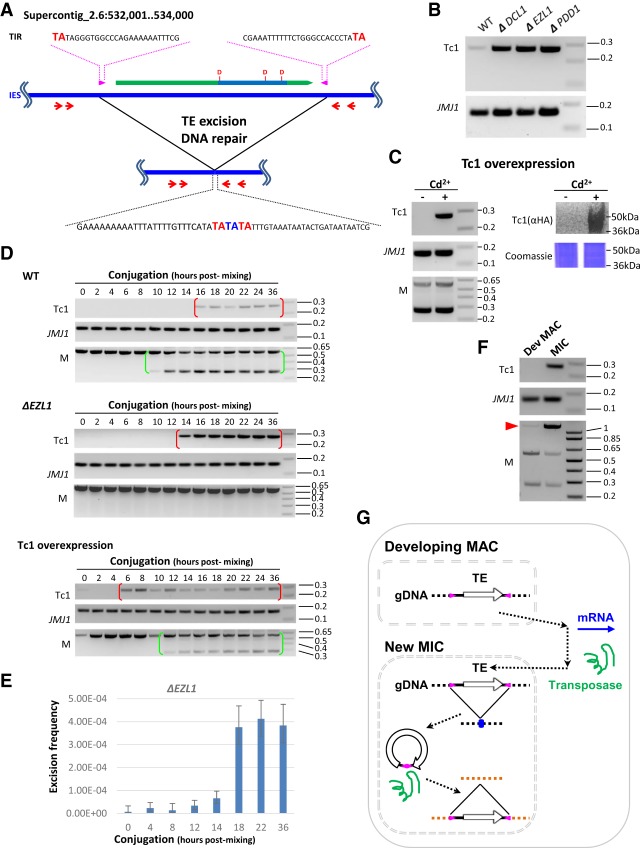
Figure 4.
Germline mobilization of a recently active TE. (A) A schematic for mobilization of a Tc1 element. It contains a conserved DNA transposase (ORF: green block arrow; transposase domain: indigo box; catalytic triad: DDD). The Tc1 element also features intact TIR (magenta arrow box) and TSD (TA in bold red in the top and bottom sequences), while its excision leaves a TE footprint (TA in bold blue in the bottom sequence). The red arrows represent the nested PCR primers used in the transposition assays shown in panels B–D. The boundaries of the IESs lie outside the PCR primers, so the somatically rearranged DNA was undetected by this assay. (B) Mobilization of the Tc1 element at low levels in wild-type cells but at dramatically increased levels in the mutants. To monitor excision at the Tc1 element, nested PCR was performed on total genomic DNA purified at the end of conjugation (24 h after mixing) from the specified cells. A MAC gene, JMJ1, was monitored as the loading control. (C) Tc1 mobilization by transposase overexpression. To monitor excision at the Tc1 element, nested PCR was performed on total genomic DNA purified at the end of conjugation (24 h after mixing), with or without induction of the Tc1 transposase overexpression by cadmium (±Cd2+). A MAC gene, JMJ1, was monitored as the loading control. Completion of conjugation was monitored by a PCR assay for a well-studied IES, the M element (Duharcourt and Yao 2002; Liu et al. 2004). Overexpression of the Tc1 transposase (HA-tagged) was shown by immunoblotting with an anti-HA antibody. Coomassie-stained total cellular proteins was used as the loading control. (D) Time course analysis of Tc1 mobilization. In wild-type cells, the Tc1 excision product was detected after developmentally programmed DNA elimination and persisted in conjugating cells (red brackets). Conjugation progress was monitored by a PCR assay for a well-studied IES, the M element. Its processing gives rise to the short PCR product in conjugating CU427 and CU428 cells (green brackets), distinct from the long PCR product from the mature MAC in parental cells (Duharcourt and Yao 2002; Liu et al. 2004). In ΔEZL1 cells, dramatically increased levels of the Tc1 excision product were detected, but with a temporal pattern similar to wild-type cells (red brackets). In cells overexpressing the Tc1 transposase, excision of the Tc1 element was expedited. JMJ1 was monitored as the loading control. (E) Excision frequency of the Tc1 element in ΔEZL1 cells. Genomic DNA samples were isolated from the specified conjugation time points, digested by HindIII to release the Tc1 element-containing IES fragment, and analyzed by Droplet Digital PCR. Excision frequency was quantified by dividing the number of Tc1 excision events with the number of control events. (F) Tc1 mobilization occurs in the new MIC. The developing MAC and the new MIC were purified from wild-type cells at late conjugation (24 h after mixing). The M element was monitored for the enrichment of MAC and MIC. The two bottom bands correspond to the IES-excised forms found only in MAC, while the top band corresponds to the IES-retained form found only in MIC (red arrowhead). The former were enriched in the developing MAC sample, while the latter was enriched in the new MIC sample. JMJ1 was the loading control. (G) A model for TE mobilization in Tetrahymena and other binucleated ciliates. (1) TE mRNA is generated in the developing MAC. (2) Transposases are imported into the new MIC. (3) TEs are mobilized in the new MIC: (3.1) excision from a donor locus, leaving behind a TE footprint (blue dot; see A); (3.2) insertion into a receptor locus.