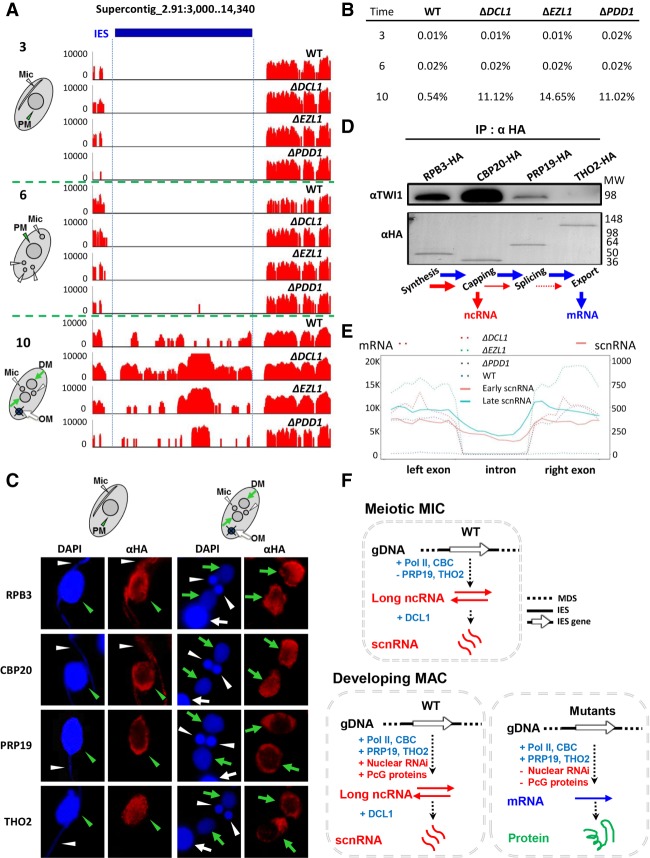
Figure 6.
The balance between ncRNA and mRNA production is a critical aspect of the balance between transcriptional silencing and activation. (A) IES-specific polyadenylated transcripts are only produced during developing MAC formation. (3) 3 h after mixing: meiosis, (6) 6 h after mixing: gametogenesis, (10) 10 h after mixing: developing MAC formation, (blue bar) IES. RNA-seq was performed after oligo-dT enrichment of polyadenylated transcripts. Note that essentially no RNA-seq reads were mapped to the IES region at the two early conjugation time points. (PM) Parental MAC, (DM) developing MAC, (OM) old MAC. (B) Percentage of RNA-seq reads mapped to consistently processed IESs, relative to total mappable reads. Note that IES-specific polyadenylated transcripts were abundantly produced during developing MAC formation (10 h after mixing) but rarely detected before that (3 and 6 h after mixing). (C) Localization of the transcriptional and cotranscriptional machineries in early (3 h after mixing) and late (10 h after mixing) conjugating cells. RPB3-HA, CBP20-HA, PRP19-HA, and THO2-HA cells were stained with an anti-HA antibody (red) and counterstained with DAPI (blue). Parental MAC (PM): green arrowhead, developing MAC (DM): green arrows, MIC: white arrowheads, old MAC (OM): white arrow. (D) Interaction between TWI1 and the transcriptional/cotranscriptional machineries. The designated cells were processed for crosslink-immunoprecipitation with the anti-HA antibody at late conjugation (10 h after mixing). The anti-HA and anti-TWI1 antibodies were used for immunoblotting. Note that similar amounts of bait proteins were recovered, as shown by the anti-HA immunoblotting. (E) Production of scnRNA from intronic as well as exonic regions. Distributions of polyadenylated transcripts (dotted lines) and scnRNA (solid lines) in introns and exons of IES-specific loci. Each intron is equally divided into 10 units from the 5′ to 3′ end; the two flanking exons are also equally divided into 10 units, respectively. The average RPM of each unit in all the loci were cumulated. Double counting for exons is avoided. (F) Alternative production of ncRNA and mRNA in the meiotic MIC and the developing MAC. In this simplified schematic, transcription from IES-specific loci is emphasized, while genic transcription from MDS is omitted. See text for details.