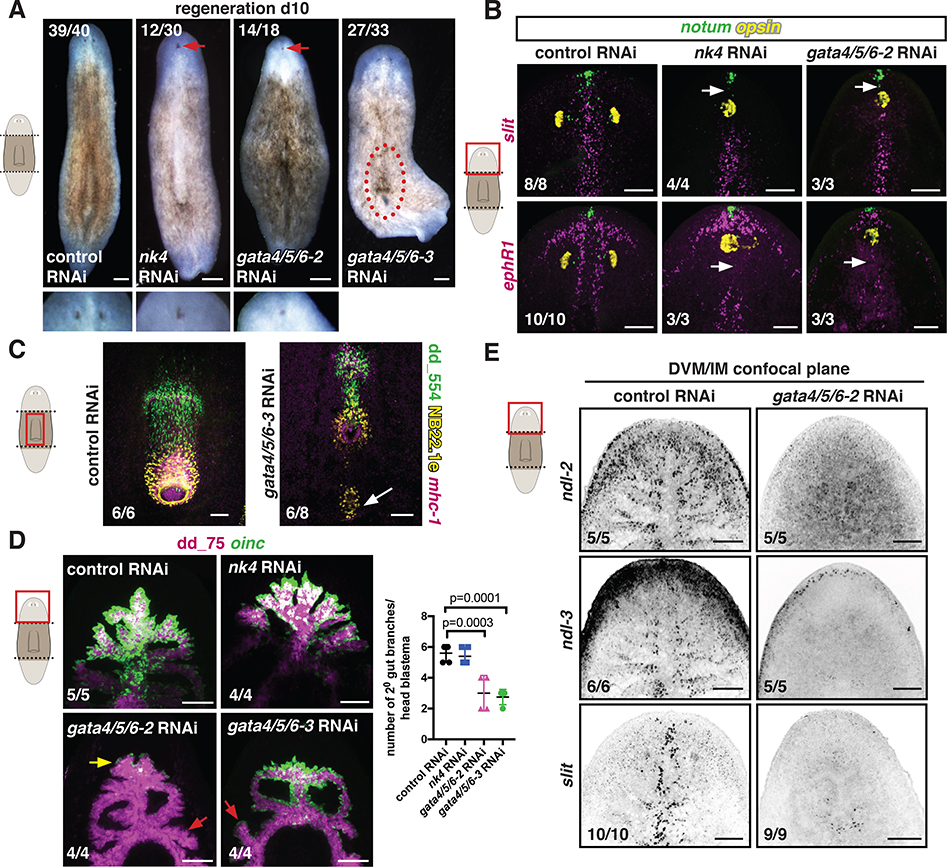
Figure 4. nk4 and gata4/5/6–2 are required for normal medial-lateral patterning during regeneration.
(A) Regenerating RNAi animals. Higher magnification shows cyclopia. Red-dotted line, ectopic pharynx. (B) Abnormal expression patterns (white arrows) of midline genes (slit, ephR41) by FISH. opsin, photoreceptor neurons; notum, anterior pole. (C) Ectopic pharynx and mouth (white arrow) in a gata4/5/6–3 RNAi animal. (D) Reduced oinc+ cells (yellow arrow) in regenerating gata4/5/6–2 RNAi animals and abnormal intestinal branches in gata4/5/6–2 and gata4/5/6–3 RNAi animals (red arrows). Right, secondary branch numbers. Mean ± SD. One-way ANOVA, post Dunnetts’ test. (E) Reduced DVM expression of PCGs in a gata4/5/6–2 RNAi animal. Images are maximal intensity projections of the entire DV axis in B-D, or of planes around intestinal branches in E. Cartoons indicate image location. Bars, 100 μm. See also Figure S6.