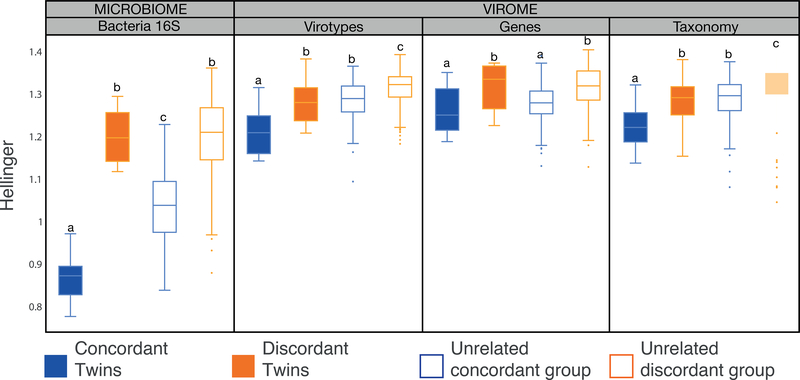
Figure 6. Virome Beta-diversity patterns mirror microbiome Beta-diversity.
Box plots show the distribution of Hellinger distances for microbiomes and viromes, according to the three different layers of information recovered (virotypes, genes, and taxonomy), for concordant co-twins (solid blue, n=9), discordant co-twins (solid orange, n=12), unrelated samples within the concordant co-twins (blue outline, n=144), and unrelated samples within the discordant co-twins (orange outline, n=264). Significant differences between means (Mann-Whitney’s U test, p < 0.020) are denoted with different letters.