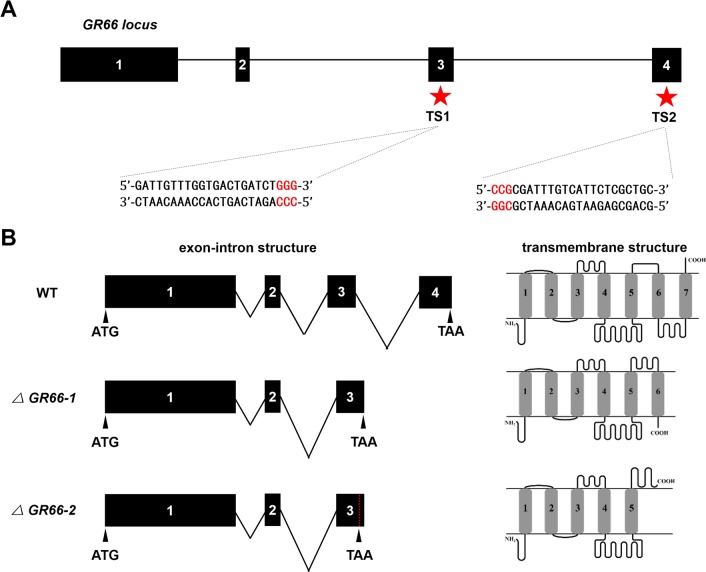
Fig 2. CRISPR/Cas9-mediated knockout and comparison among WT and mutants.
(A) Schematic depiction of the GR66 locus and sgRNA targeting sites. The sgRNA targeting sites, namely, TS1 and TS2, are located on the sense strand of exon-3 and the antisense strand of exon-4, respectively. The sgRNA targeting sequence is shown in black, and the PAM sequence is shown in red. (B) Comparison of gene structure among the WT and two homozygous mutant lines. Left, exon-intron structure of GR66. The 3ʹ fragment of exon 3, the third intron and the 5ʹ fragment of exon 4 were excised in both homozygous mutant lines. In ΔGR66-2, excision of the sequences caused frameshift mutations. The dotted red line indicates the premature termination codon. Right, transmembrane domain predictions of GR66 for the WT and two homozygous mutant lines. In WT, the GR66 protein consists of seven transmembrane domains, an intracellular N terminus and an extracellular C terminus. In ΔGR66-1, the truncated protein consists of six transmembrane domains, and the N terminus and C terminus are both extracellular. In ΔGR66-2, the truncated protein consists of only five transmembrane domains. The orientation of the N terminus and C terminus are the same in WT. CRISPR/Cas9, clustered regularly interspaced short palindromic repeats/CRISPR-associated protein-9 nuclease; GR66, gustatory receptor 66; PAM, protospacer adjacent motif; sgRNA, small guide RNA; WT, wild type.