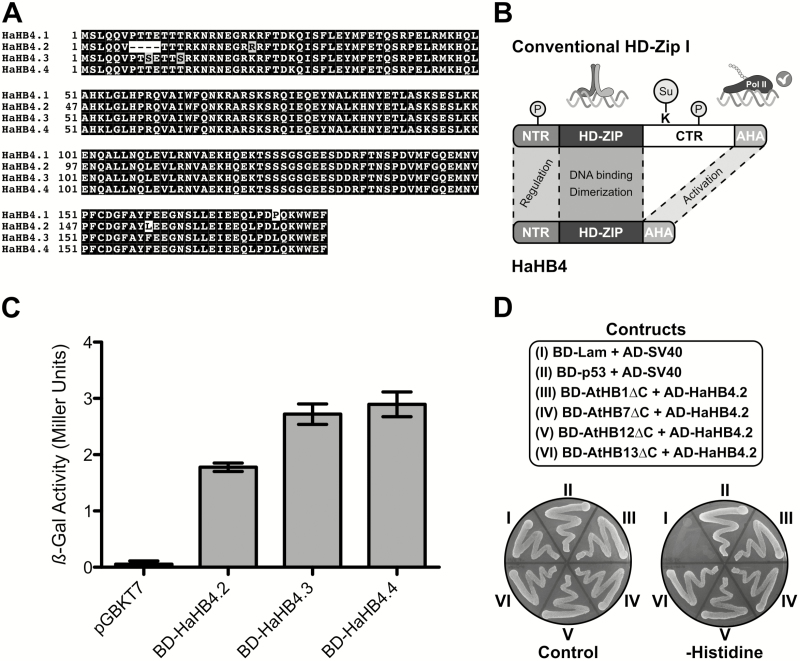
Fig. 1.
HaHB4.2 binds HD-Zip I transcription factors and activates transcription in yeast. (A) Multiple sequence alignment of the different HaHB4 isoforms. Darker background corresponds to enhanced conservation and numbers highlight the positions of amino acids. HaHB4.1 represents the WT form of sunflower HaHB4. (B) Schematic representation of the domains exhibited by most HD-Zip I and HaHB4. AHA, aromatic and large hydrophobic residues embedded in an acidic context; CTR, carboxy-terminal region; HD-Zip, homeodomain-leucine zipper; NTR, amino-terminal region; P, phosphorylation; Su, SUMOylation. (C) HaHB4 isoforms act as transcriptional activators in yeast-one hybrid assays. β-Galactosidase activity was quantified in Miller units and error bars represent standard deviation of three independent technical triplicates. (D) HaHB4.2 interacts with HD-Zip I from Arabidopsis. Yeast two-hybrid analysis of HaHB4.2 (as a Gal4-activating domain fusion; AD) with AtHB1, AtHB7, AtHB12, and AtHB13 lacking the CTR as a Gal4-DNA-binding domain (BD) fusion). Growth on plates without histidine indicates protein interaction. SV40 large-T antigen (AD-SV40) with either human lamin C (BD-Lam) or murine p53 (BD-p53) was used as negative and positive controls, respectively.