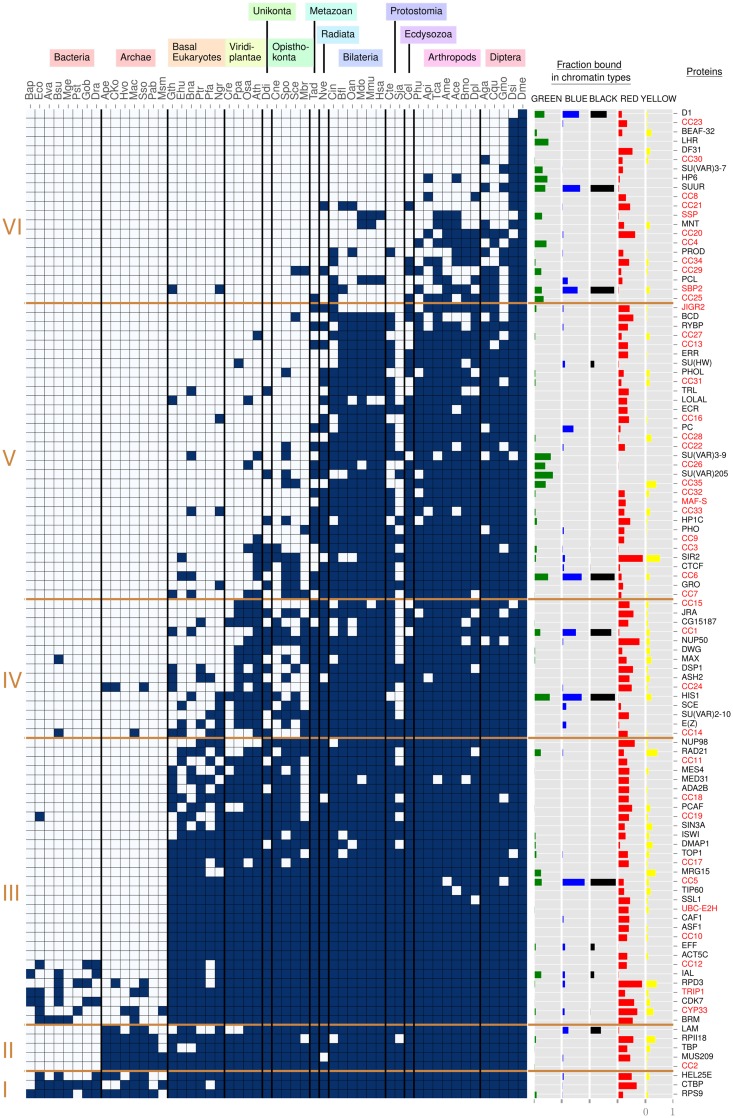
Fig. 1.
—Phylogenetic profile of chromatin-associated proteins. To the left, six protein age clusters resulting from clustering with partitioning around medoids (see Materials and Methods for details). They are indicated with Roman numerals (I–VI). On top, 13 species groups are manually defined to aid the reader, three letter codes refer to species names as given in supplementary table 1, Supplementary Material online. In the matrix, dark blue rectangles represent the presence of a homolog, gray rectangles its absence. Within each age cluster, rows are ordered from top to bottom by decreasing number of dark blue rectangles. Columns are ordered at the level of species groups by decreasing phylogenetic distance to Drosophila melanogaster, with Drosophila (Dme) in the rightmost column (see supplementary fig. 1, Supplementary Material online, for details). Within a species group, columns are arbitrarily ordered. The five columns “Fraction bound in chromatin types” display the fraction of chromatin type (GREEN, BLUE, BLACK, RED, YELLOW) bound by each CAP. To the right, the column “Proteins” contains protein names, with unknown proteins in a red font.