Fig. 6.
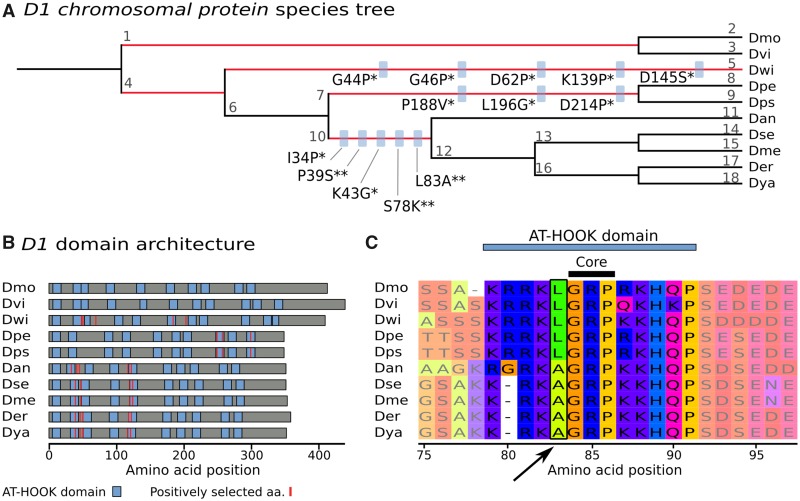
—D1 Chromosomal protein is undergoing positive selection at AT-HOOK domains. (A) Drosophilid species tree (with arbitrary branch lengths). The five branches with positive selection events are highlighted in red (P < 0.01, Bonferroni correction). On branches with more than one positively selected site, blue boxes indicate the amino acid substitution under positive selection, with the significance given as posterior probability of dN/dS >1 (* for Pr >0.95, ** for Pr >0.99). For instance, L83A indicates the substitution of lysine with alanine at position 83. The Drosophila species are indicated by three letter abbreviations: Dmo is D. mojavensis, Dvi is D. virilis, Dwi is D. willistoni, Dpe is D. persimilis, Dps is D. pseudoobscura, Dan is D. ananassae, Dse is D. sechellia, Dme is D. melanogaster, Der is D. erecta, and Dya is D. yakuba. Each branch is identified with a number that links to details on positive selection tests (supplementary table 7, Supplementary Material online). (B) Domain architecture of D1 proteins. Per species, protein length is given by gray horizontal bars. AT-HOOK domains are represented by blue boxes and positively selected amino acids (aa) are displayed as red vertical lines. (C) An example of positive selection in an AT-HOOK domain. The multiple alignment of D1 is zoomed in on the region 75–97aa, showing a change of L83 directly in front of the core motif. Visualization is done with MSAViewer (http://msa.biojs.net/app/; last accessed February 6, 2019).