FIGURE 3.
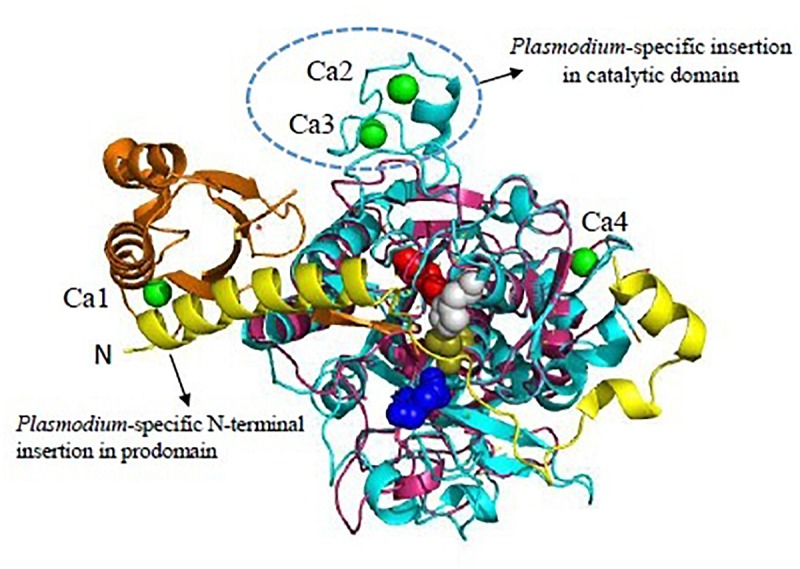
Unique features of Plasmodium vivax SUB1 structure. Cartoon representation of overall structure of PvSUB1 (PDB: 4TR2). Overall structure comprises N-terminal insertion in prodomain comprising the “belt” domain (shown in yellow) followed by a classical bacterial-like prodomain (orange), and the C-terminal subtilisin-like catalytic domain (cyan). The catalytic triad (Asp316/His372/Ser549; Red/Gray/olive) is shown as spheres. The oxyanion hole residue (Asn464) is shown as blue sphere. Bound calcium ions are shown as green spheres. Out of four calcium binding sites, three (Ca1, Ca2, Ca3) are specific to parasite subtilases. Ca4 is conserved in all homologs of subtilisin. Bacillus subtilsin (PDB: 1SUP) is superimposed (shown in magenta) highlighting the unique insertions in parasite subtilases. All structure figures have been made in Pymol.
