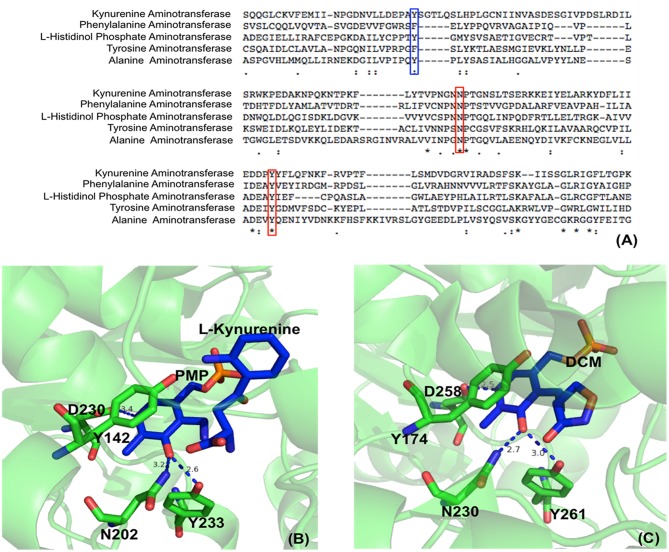
Figure 2.
Partial protein sequence alignment of aminotransferases, and the position of three conserved residues in the active site. Sequences of Homo sapiens kynurenine aminotransferase (Accession: NP_057312.1), Mycobacterium tuberculosis H37Rv phenylalanine aminotransferase (PDB: 4R2N, chain A) (Nasir et al., 2016), Enterobacteriaceae L-histidinol phosphate aminotransferase (Accession: WP_000108941.1), Homo sapiens tyrosine aminotransferase (PDB: 3DYD, chain A) (http://www.rcsb.org/structure/3DYD) and Hordeum vulgare alanine aminotransferase (PDB: 3TCM, chain A) (Duff et al., 2012) are shown. The uncharged aromatic amino acid residues for π-π stacking with PLP ring are highlighted in blue box while the Asn and Tyr residues forming hydrogen bonds with PLP O3′ are highlighted in red boxes (A). The Asn and Tyr residues forming hydrogen bonds with O3′ of PLP are shown in green sticks while the ligands pyridoxamine phosphate (PMP), L-kynurenine and D-pyridoxyl-N,O-cycloserylamide-5-monophosphate (DCM) are shown in blue sticks. The uncharged aromatic amino acid residues forming π-π stacking with PLP pyridine ring and the Asp residue near pyridine ring N atom are also shown in green sticks. The distances between PLP O3′ and Asn or Tyr and the distances between pyridine ring N and Asp side chain are in blue dashed line and labeled (Unit: Å). L-kynurenine aminotransferase (PDB: 2R2N) (Han et al., 2008b) (B) and alanine aminotransferase (PDB: 3TCM) (Duff et al., 2012) (C) are presented as two examples.