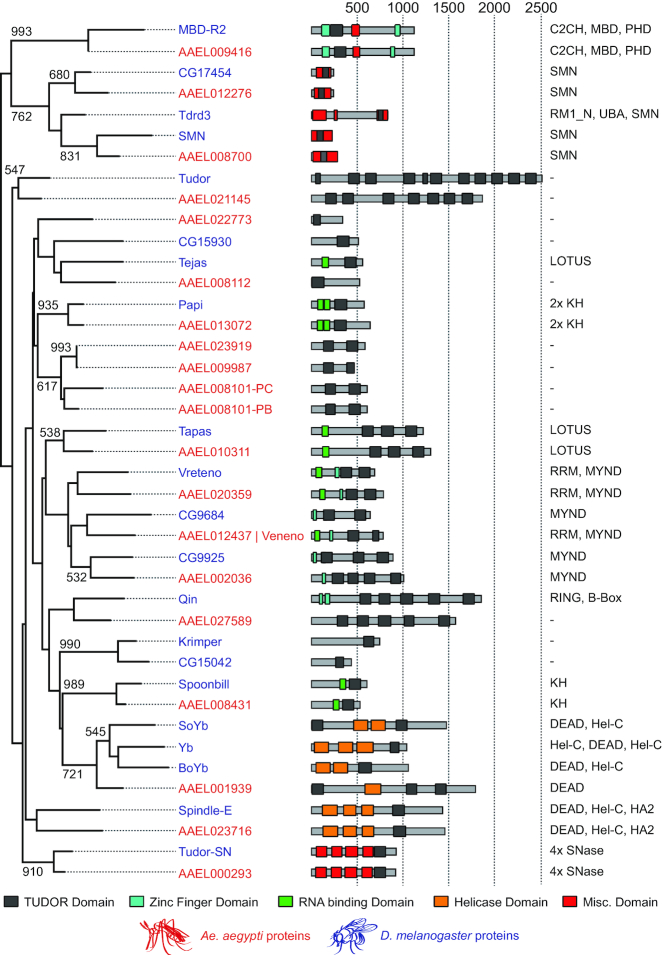
Figure 1.
Orthologous Tudor genes in Drosophila melanogaster and Aedes aegypti. On the left, a neighbor joining tree based on TUDOR domains from A.aegypti (red) and D. melanogaster (blue) is shown. Numbers indicate bootstrap values for 1000 iterations; only values >500 are shown. In the middle, predicted domain structures of Tudor proteins are drawn schematically, with TUDOR domains shown in black, zinc fingers in blue, putative RNA binding domains in green, domains associated with helicase activity in orange and all other domains in red. Numbers at the top indicate protein length in amino acids. On the right, protein domains other than TUDOR domains are presented, ordered from amino to carboxyl terminus, as indicated in the middle panel. As the AAEL008101 gene produces two splice variants encoding TUDOR domains of slightly different composition (PB and PC), both were included as separate entities in the multiple sequence alignment. B-box, B-box type zinc finger (Zf) domain; C2CH, C2CH-type Zf domain; C2H2, C2H2-type Zf domain; DEAD, DEAD box domain; HA2, Helicase-associated domain; Hel-C, helicase C domain; KH, K homology RNA-binding domain; LOTUS, OST-HTH/LOTUS domain; MBD, Methyl-CpG-binding domain; MYND, MYND (myeloid, Nervy, DEAF-1)-type Zf domain; PHD, PHD-type Zf domain; RING, RING-type Zf domain; RMI1_N, RecQ mediated genome instability domain; RRM, RNA recognition motif; SMN, survival motor neuron domain; SNase¸ Staphylococcal nuclease homologue domain; UBA, ubiquitin associated domain.