Abstract
In the promoter of c-KIT proto-oncogene, whose deregulation has been implicated in many cancers, three G-rich regions (kit1, kit* and kit2) are able to fold into G-quadruplexes. While kit1 and kit2 have been studied in depth, little information is available on kit* folding behavior despite its key role in regulation of c-KIT transcription. Notably, kit* contains consensus sites for SP1 and AP2 transcription factors. Herein, a set of complementary spectroscopic and biophysical methods reveals that kit*, d[GGCGAGGAGGGGCGTGGCCGGC], adopts a chair type antiparallel G-quadruplex with two G-quartets at physiological relevant concentrations of KCl. Heterogeneous ensemble of structures is observed in the presence of Na+ and NH4+ ions, which however stabilize pre-folded structure. In the presence of K+ ions stacking interactions of adenine and thymine residues on the top G-quartet contribute to structural stability together with a G10•C18 base pair and a fold-back motif of the five residues at the 3′-terminal under the bottom G-quartet. The 3′-tail enables formation of a bimolecular pre-folded structure that drives folding of kit* into a single G-quadruplex. Intriguingly, kinetics of kit* G-quadruplex formation matches timescale of transcriptional processes and might demonstrate interplay of kinetic and thermodynamic factors for understanding regulation of c-KIT proto-oncogene expression.
INTRODUCTION
The c-KIT proto-oncogene encodes a transmembrane tyrosine kinase receptor (c-kit) (1,2), which participates, once activated by endogenous ligands, in a broad range of physiological processes, including cell proliferation, migration, maturation and survival (3). The overexpression and/or mutations of c-KIT gene have been implicated in a number of human cancers, like gastrointestinal stromal tumors, pancreatic cancer, leukemia and melanoma (4,5). In the proximal promoter of c-KIT three guanine (G)-rich regions have been identified that are able to fold into G-quadruplexes (3,6,7). In general, G-quadruplexes consist of stacked G-quartets, cyclic arrangements of four guanine residues held together by Hoogsteen hydrogen bonds and stabilized by a central cation (8). The G-quadruplex structures are highly polymorphic and are influenced by factors such as oligonucleotide sequence and concentration, type and concentration of cations in solution, crowding conditions and presence of other biomolecules (9–14). In general, under in vitro conditions G-quadruplexes adopt parallel, antiparallel and hybrid (3+1) topologies characterized by different orientations of the four G-rich tracts. Three types of loops that connect guanine residues involved in G-quartets are typical for G-quadruplexes and can adopt propeller, diagonal and edgewise orientations (15). G-quadruplexes have been clearly shown to exist inside human cells (16–19). Moreover, bioinformatic analysis revealed that G-rich repeats are not randomly distributed in the human genome, but are mostly located in regions associated with a number of essential biological processes such as transcription, replication and telomere maintenance (20,21). In addition to c-KIT proto-oncogene, more than 40% of genes that encode human proteins contain one or more putative G-quadruplex forming motifs in the promoter regions (22). Stabilization of G-quadruplex structures is a novel promising strategy to regulate gene expression at transcriptional and translational levels (23–27). For example, stabilization of G-quadruplex structures by small molecule ligands in the promoter region of c-KIT gene has been linked with inhibition of its transcriptional activity and reduction of the expression of c-kit tyrosine kinase receptor which possibly lead to favorable anticancer effects (28–30).
The G-quadruplex-forming regions within the c-KIT promoter have been named kit1, kit* and kit2 (3,6,7). They are closely clustered and separated from each other by only few nucleotides (six between kit1 and kit*, and only one between kit* and kit2, Figure 1A). kit1 and kit2 oligonucleotides contain four GGG-repeats each that fold into parallel G-quadruplexes with three G-quartets under in vitro conditions. Their structural characteristics as well as thermodynamic behavior have been extensively studied (6,31–38). Worth of note, kit1 G-quadruplex contains unique structural features; an isolated guanine that is involved in the formation of the G-quartet core and a stem-loop consisting of 5 nt (32). In the case of kit2, complex folding pathway was observed involving stable folding intermediates that convert into thermodynamically stable monomeric and dimeric parallel G-quadruplex structures (33,34). High-resolution structure of kit* is not available. It is noteworthy that kit* contains consensus sites for SP1 and AP2 transcription factors (3,7,39). Therefore, its duplex–quadruplex equilibrium might modulate c-KIT transcription. Additionally, we have demonstrated recently that cross-talking interactions stabilize kit* and kit2 G-quadruplexes and impair their refolding into duplex form in the presence of the complementary strands (40). We deemed it essential to get deeper insights into structural details of kit* G-quadruplex.
Figure 1.
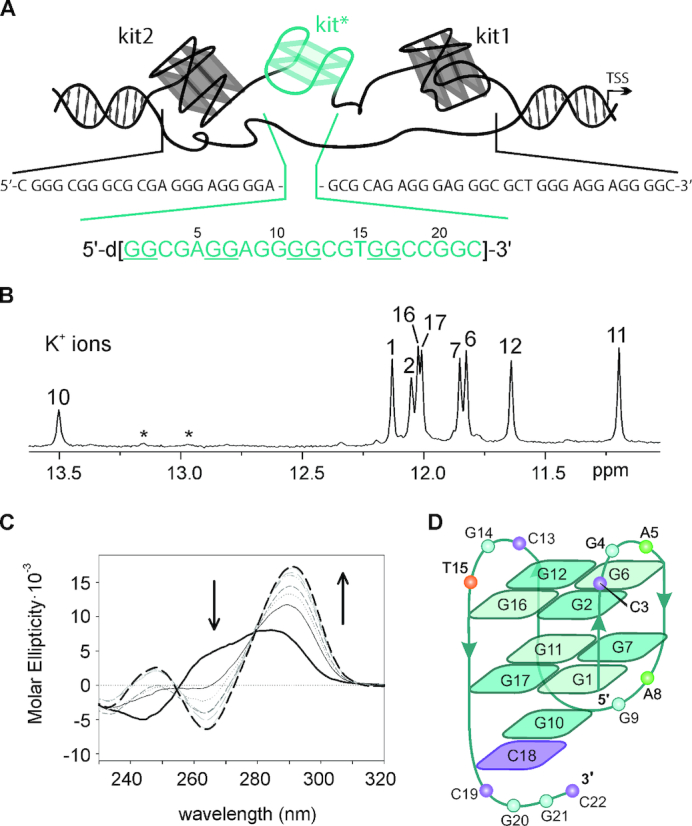
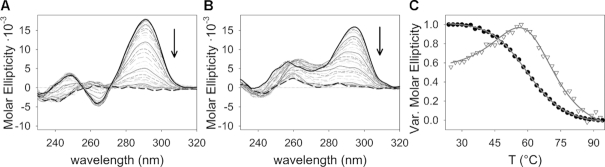
Characterization of a G-quadruplex adopted by kit*. (A) Schematic representation of G-rich region in the promoter of c-KIT gene and its primary sequence. The arrow indicates the transcription start site. (B) Imino region of 1H NMR spectrum of kit* in the presence of 100 mM KCl, 0.5 mM kit* concentration per strand, pH 7.4 and 37°C on an 800 MHz NMR spectrometer. Assignments are shown above the individual signals. Signals marked with asterisks correspond to pre-kit*. (C) CD spectra of kit* at 4 μM concentration per strand titrated with KCl in 10 mM TRIS, pH 7.5 at 25°C. Arrows indicate changes in CD spectra from 0 mM (black solid line) to 350 mM KCl (black dashed line). (D) Topology of G-quadruplex adopted by kit*. Anti and syn guanines in G-quartets are marked with darker and lighter shades of cyan, respectively.
Herein, we describe the results of our studies on structural features of kit*, d[GG CGA GG AGG GG CGT GG CCGGC], performed by using a set of complementary spectroscopic and biophysical methods. A kit* sequence containing a GGGG-tract as well as four GG-repeats could be expected to fold into multiple G-quadruplex structures. In contrast, kit* adopts a single antiparallel two G-quartet G-quadruplex at physiologically relevant temperature (37°C) and concentration of K+ ions (100 mM) as revealed by nuclear magnetic resonance (NMR). Loop regions consisting of three regions of three residues, each are prone to interact with each other as well as with the 3′-tail and thus contribute to the stabilization of the structure. Two adenine residues in the loop regions provide excellent opportunity to evaluate their stacking interactions or exposure to solvent with fluorescence spectroscopy. Searching for a rationale for the observed conformational selection of kit* uncovered importance of the last five residues at the 3′-terminal. Shortening of the 3′-tail disturbed its ability to promote formation of a dimeric assembly in the absence of K+ ions and was expected to reveal a novel pre-folded structure that drives folding into a single G-quadruplex. We deemed it essential to explore folding behavior of kit*. Upon addition of K+ ions, kit* folds into a G-quadruplex within seconds, but if the 3′-tail is absent folding pathway changes and many parallel G-quadruplexes are formed. Interesting interplay of kinetic and thermodynamic factors orchestrated by the 3′-tail uncovered its crucial role in folding process, thermal stabilization and structural integrity of kit* G-quadruplex thus alike contributing to fine regulation of c-KIT proto-oncogene expression in physiological conditions.
MATERIALS AND METHODS
NMR sample preparation
The isotopically unlabeled and residue-specific partially 15N, 13C-labeled (10% guanine and 4% cytosine residues) kit* and modified oligonucleotides (Table 1) were synthesized and prepared as described before (41,42). Oligonucleotides were purified and desalted with the use of Amicon Ultra-15 Centrfugal Filter Units to give NMR samples with concentration between 0.1 and 2.0 mM per strand. NMR samples of kit* and modified oligonucleotides were prepared by dissolving desalted oligonucleotides in 300 μl of 90%/10% mixture of H2O/2H2O in 20 mM potassium phosphate buffer (pH 7.4) or in 20 mM lithium cacodylate buffer (pH 7.2) and in the presence of KCl with varying concentrations from 10 to 250 mM as well as in 100 mM concentrations of LiCl, NaCl, NH4Cl and KCl. For samples dissolved in 100% 2H2O we lyophilized solutions of oligonucleotides, KCl and buffer separately and subsequently dissolved and mixed them together in 300 μl 100% 2H2O.
Table 1.
Sequences of kit* and its analogs
| Oligonucleotide# | ||||||||
|---|---|---|---|---|---|---|---|---|
| kit* ##§ | GG | CGA | GG | AGG | GG | CGT | GG | CCGGC |
| kit*17## | GG | CGA | GG | AGG | GG | CGT | GG | |
| kit*18## | GG | CGA | GG | AGG | GG | CGT | GG | C |
| kit*19## | GG | CGA | GG | AGG | GG | CGT | GG | CC |
| kit*20 | GG | CGA | GG | AGG | GG | CGT | GG | CCG |
| kit*18T | GG | CGA | GG | AGG | GG | CGT | GG | CT |
| Tkit*18 | TGG | CGA | GG | AGG | GG | CGT | GG | C |
| Akit*18 | AGG | CGA | GG | AGG | GG | CGT | GG | C |
| kit*19TTT | GG | CGA | GG | AGG | GG | CGT | GG | CCTTT |
| kit*T15A | GG | CGA | GG | AGG | GG | CGA | GG | CCGGC |
| kit*A5T | GG | CGT | GG | AGG | GG | CGT | GG | CCGGC |
| kit*A8T | CGA | GG | TGG | GG | CGT | GG | CCGGC | |
| kit*C18,19T | GG | CGA | GG | AGG | GG | CGT | GG | TTGGC |
| kit*C19T | GG | CGA | GG | AGG | GG | CGT | GG | CTGGC |
| kit*C18T | GG | CGA | GG | AGG | GG | CGT | GG | TCGGC |
| kit*C13T | GG | CGA | GG | AGG | GG | TGT | GG | CCGGC |
| kit*C3T | GG | TGA | GG | AGG | GG | CGT | GG | CCGGC |
| Tkit* | TGG | CGA | GG | AGG | GG | CGT | GG | CCGGC |
| Akit* | AGG | CGA | GG | AGG | GG | CGT | GG | CCGGC |
| c | GC | CGG | ||||||
| kit*5–2AP### | GG | CG-2AP | GG | AGG | GG | CGT | GG | CCGGC |
| kit*8–2AP### | GG | CGA | GG | 2AP-GG | GG | CGT | GG | CCGGC |
#Sequences are reported in the 5′-3′ direction. Guanine residues involved in G-quartets of kit* are underlined. ## Melting temperatures are 60.5 ± 0.1°C (TRIS buffer) and 55.6 ± 0.1°C (K-phosphate buffer) for kit*, 50.6 ± 0.6°C, 75.9 ± 0.1°C (TRIS buffer) and 48.8 ± 0.6°C, 65.1 ± 0.6°C (K-phosphate buffer) for kit*17, 45.7 ± 0.4°C, 58.1 ± 0.5°C (K-phosphate buffer) for kit*18, 52.7 ± 0.1°C (K-phosphate buffer) for kit*19. ### 2AP stands for 2-aminopurine, a fluorescent analog of adenine. §Analogs of kit* with single substitution of dG with 8Br-dG at positions 1, 6, 11 and 16 are presented in Supplementary Figure S20.
NMR experiments
NMR data were collected on Agilent (Varian) NMR System 600 and 800 MHz spectrometers equipped with triple-resonance HCN cryogenic and PFG One NMR probes in the temperature range from 5 to 65°C. The majority of NMR spectra were recorded at 37°C with samples dissolved in 90%/10% H2O/2H2O. DPFGSE_NOESY spectra were acquired at mixing times of 40, 80, 150, 200, 300 and 450 ms. 13C- and 15N-edited HSQC experiments were recorded on residue specifically 15N, 13C-labeled oligonucleotides. DQF-COSY, TOCSY (τm 40 and 80 ms), 1H–31P COSY and 1D 31P NMR spectra NMR spectra were acquired on NMR samples prepared in 100% 2H2O solution. NMR spectra were processed and analyzed by using VNMRJ (Varian Inc.) and the Sparky (UCSF) software. Cross-peak assignment and integration was achieved using Sparky (UCSF) software.
Circular dichroism (CD) spectroscopy
Circular dichroism (CD) spectra were acquired on a JASCO J-810 spectropolarimeter equipped with a Peltier temperature controller. CD spectra were recorded from 230 to 320 nm with the following parameters: scanning speed of 100 nm/min, band width of 2 nm, data interval of 0.5 nm and response of 2 s. Measurements were performed using a 1 cm path length quartz cuvette at oligonucleotide concentration of ∼4 μM in 10 mM TRIS (pH 7.5). CD titrations with KCl or LiCl were performed at 25°C. After each titration step the system was left to equilibrate before spectra acquisition. CD signal variations as a function of the concentration of cations at the maximum were fitted by applying one-site saturation model (Supplementary Data, Equation 1). For CD kinetic experiments, KCl was added manually to the cuvette from a stock solution and mixing was provided by an in-cuvette magnetic stirring bar. After a mixing time of 10 s spectra acquisition was initiated using an interval scan of 30 min. The temperature was maintained at 37°C (Supplementary Data, Equation 2). CD melting studies were performed on kit* and kit*17. The experiments were carried out between 25 and 95°C in 150 mM KCl. The heating rate was 50°C/h, each 2°C the temperature was held for 5 min, the corresponding CD signals were recorded at 294 and 264 nm, in the case of kit* and kit*17, respectively, and fitted according to van′t Hoff formalism (Supplementary Data, Equations 3 and 4).
Fluorescence spectroscopy
Fluorescence measurements were performed on kit*5–2AP and kit*8–2AP by using a JASCO-FP-6500 spectrofluorometer equipped with a Peltier Temperature controller. Emission spectra were recorded in the 320–460 nm range with the excitation wavelength fixed at 305 nm. The scanning speed was 200 nm/min, the response was 1 s and both the emission and the excitation bandwidths were 3 nm. The fluorescently labeled oligonucleotides were diluted to a concentration of 0.4 μM in 10 mM TRIS (pH 7.5). The effect of reaching 150 mM KCl on the fluorescence spectra was monitored at 25°C. Acrylamide quenching experiments were performed on previously folded oligonucleotides in 150 mM KCl. The quenching efficiency was monitored at 370 nm at 25°C and plotted as a function of the acrylamide concentration. Data points were fitted according to Stern–Volmer formalism to obtain quenching parameters (Supplementary Data, Equations 5 and 6).
Restraints, structure calculations and molecular dynamics simulations
For structural calculations we used NOE-derived distance (force constant 20 kcal mol−1 Å−2), hydrogen bonds (force constant 20 kcal mol−1 Å−2) and torsion angles χ (force constant 200 kcal mol−1 rad−2) restraints. The average volume of H6-Me(H7)-cross-peak of T15 was used as the distance reference of 3.0 Å. Cross-peaks were classified as strong (1.8–3.6 Å), medium (2.5–5.0 Å), weak (3.5–6.5 Å) and very weak (4.5–7.5 Å). NOE distance restraints for protons were obtained from a 2D NOESY spectrum recorded with mixing time of 150 ms. Some additional cross-peaks of weak intensities were gathered at mixing times up to 450 ms and classified as very weak cross-peaks. The torsion angle χ restraints were applied for all of residues based on the intensity of respective H8-H1′ 2D NOESY cross-peaks, (for region 200–280° for adenines and guanines with anti-orientation along glycosidic bonds, 25–95° for guanines with syn-orientation along glycosidic bonds, 170–310° for thymine and cytosine residues). The structures of kit* G-quadruplex were calculated by the simulated annealing simulations. The force field parameters were adopted from the Generalized Amber force field (43) (Supplementary Data). For molecular dynamics (MDs) simulations, the kit* G-quadruplex was placed in a truncated octahedral box of TIP3P water molecules with the box border at least 10 Å away from any atoms of the G-quadruplex. Extra K+ ions were added to neutralize the negative charges of the G-quadruplex. Prior to MD simulations, the system was subjected to a series of minimizations and equilibrations (Supplementary Data).
RESULTS
K+ ions induce folding of kit* into a well-defined G-quadruplex, while Li+, Na+ and NH4+ ions stabilize pre-folded structure through GC base pairs
1H NMR spectrum of kit* in the presence of 100 mM LiCl at 37°C exhibits two broad signals at δ 12.94 and 13.14 ppm that are typical for imino protons of guanine residues involved in GC base pair formation (Supplementary Figure S1). No imino signals are detected in the region between δ 10.5 and 12.5 ppm which would be characteristic for imino protons of guanine residues involved in G-quartets. Thus, the two signals arise from formation of a pre-folded structure of kit* (pre-kit*) that does not contain stacked G-quartets (vide infra). In the presence of 100 mM NH4Cl at 37°C similar NMR fingerprint of imino signals has been detected for kit* as in the presence of LiCl suggesting formation of pre-kit*, and absence of G-quadruplex structure (Supplementary Figure S1). Pre-kit* is predominant species also in the presence of 100 mM NaCl at 37°C. However, we also observe eight weak and broad imino signals between δ 11.1 and 12.2 ppm in 1H NMR spectrum at 100 mM NaCl suggesting a minor formation of G-quadruplex (Supplementary Figure S1). In contrast, 1H NMR spectrum of kit* in the presence of 100 mM KCl at 37°C exhibits eight narrow and well-resolved signals between δ 11.20 and 12.13 ppm, which are consistent with formation of a single G-quadruplex with two G-quartets (Figure 1B and Supplementary Figure S1). The additional signal at δ 13.50 ppm suggests the presence of a GC base pair in Watson–Crick geometry. Interestingly, in the presence of K+ ions we also detected two broad signals at δ 12.94 and 13.14 ppm of lower intensity (∼10%) corresponding to pre-kit* that are observed in LiCl, NH4Cl and NaCl containing solutions (Supplementary Figures S1–3). The coexistence of the two distinct species in the presence of the K+ ions has been confirmed by DOSY NMR experiment at 5°C displaying two different translational diffusion coefficients of 0.87 × 10 −10 (kit* G-quadruplex) and 0.45 × 10 −10 m2 s−1 (pre-kit*) (Supplementary Figure S4). In full agreement, analysis of kit* electrophoretic mobility showed co-presence of two species, monomeric G-quadruplex and dimeric pre-kit* (Supplementary Figure S5). Despite the observation of minor species in solution, kit* G-quadruplex is predominantly favored structure in the presence of K+ ions.
Titration of kit* with KCl (0–250 mM) revealed nine (8 + 1) resolved imino 1H NMR signals (Figure 1B and Supplementary Figure S6). By increasing K+ ion concentration the intensity of imino signals increased and reached a plateau at 50 mM KCl, while no new imino signals appeared in 1H NMR spectra. The CD spectrum of kit* in the presence of KCl suggests formation of an antiparallel topology with the minimum and maximum at 264 and 294 nm, respectively (Figure 1C and Supplementary Figure S7). Isodichroic points at 255 and 279 nm are conserved between 0 and 350 mM KCl indicating that the initial folded state(s) interconvert into the final antiparallel form without participation of stable kinetic intermediates. Analysis of CD signal variation at 294 nm as a function of KCl concentration provided a dissociation constant (KD) of 14.7 ± 1.0 mM for K+ ions at 25°C (Equation 1). Furthermore, in 1H NMR spectra no significant variation of the signals associated with changes in kit* G-quadruplex structure was observed at concentrations of kit* between 0.1 and 1.7 mM (Supplementary Figure S8). The above results indicate that kit* forms a monomeric G-quadruplex. At concentrations of kit* above 1.7 mM per strand, we detected a broad and unresolved hump in the imino region of 1H NMR spectra that might be related to formation of higher order structures (Supplementary Figure S9). The intermolecular association is observed also after overnight annealing (95→25°C) at 1.0 mM and higher concentrations of kit* per strand. Noteworthy, after keeping NMR samples (0.4 mM kit* per strand) for few months at room temperature we observed decrease of signals of kit* G-quadruplex and significant increase of signals at δ 12.94 and 13.14 ppm associated with GC base pairs involving G20 and G21.
An antiparallel G-quadruplex with a two G-quartet core, three edgewise loops and a 3′-tail
Residue-specific, partial 13C,15N-labeling of guanine (10%) and cytosine (4%) residues enabled unambiguous assignment of H1 and H8 as well as H6 proton resonances of guanines and cytosines, respectively (Supplementary Figures S10–14). H8 and H2 protons of A5 as well as A8 were assigned by their substitution with thymine residues in kit*A5T and kit*A8T, which resulted in minimal changes in 1H NMR spectra with respect to kit* with exception of signals for A5/T5 and A8/T8, respectively (Supplementary Figure S15). The assignment of A5 and A8 was confirmed with the help of characteristic heteronuclear correlations involving C8 and C2 atoms in 1H-13C HSQC spectra of kit* (Supplementary Figure S16). The spectral assignment of resonances of kit* were completed with the use of through-bond (DQF-COSY and TOCSY experiments) and through-space (NOESY) correlations (Figure 2; Supplementary Figures S17–21 and Table S1).
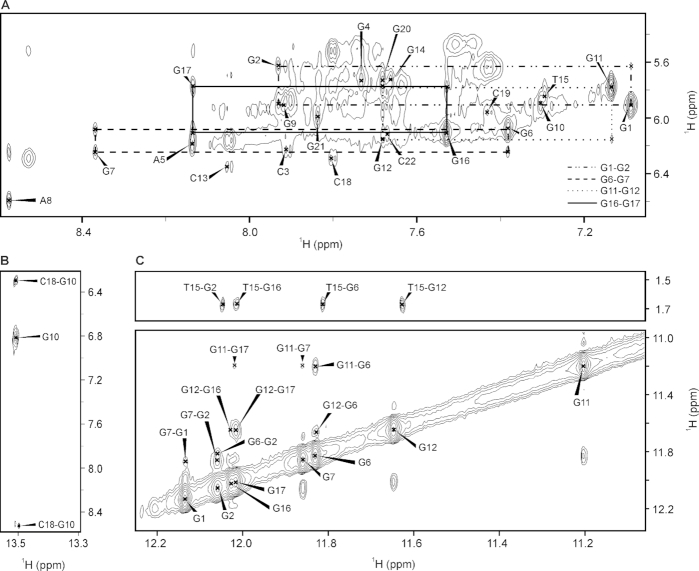
Figure 2.
Expansions of NOESY spectrum (τm 450 ms) of kit*. (A) The aromatic-anomeric region with intra-residue NOE cross-peaks marked with residue numbers. For clarity, only sequential NOE cross-peaks of G1-G2, G6-G7, G11-G12 and G16-G17, characteristic for 5′-syn-anti-3′ steps, are marked with different line styles. (B) NOE cross-peaks between G10 imino and C18 amino protons. (C) Imino-methyl and imino-imino regions. The NMR spectrum was recorded at 0.5 mM kit* concentration per strand, 100 mM KCl, pH 7.4 and 37°C on an 800 MHz NMR spectrometer.
The characteristic H1-H8 connectivities in NOESY spectra allowed us to establish topology of kit* G-quadruplex involving two G-quartets with the following hydrogen-bond directionalities: G1→G17→G11→G7 and G2→G6→G12→G16 (Figure 1D and Supplementary Figure S18). The guanine residues of G1-G17-G11-G7 quartet exhibit a clockwise donor–acceptor hydrogen-bonding directionality, while those of G2-G6-G12-G16 quartet display an anti-clockwise directionality. Orientation of G-quartets that are stacked one above the other is additionally supported by inter-quartet H1-H1 NOE cross-peaks of G2-G7, G6-G11 and G12-G17 residues (Figure 2C). Four distinct and strong cross-peaks observed in the H8-H1′ region of NOESY spectrum indicate that G1, G6, G11 and G16 residues predominantly adopt a syn conformation along glycosidic torsion angles (Supplementary Figure S20). The ‘rectangular’ pattern of NOE cross-peaks of H1′(n)-H8(n + 1)-H1′(n + 1)-H8(n) for G1-G2, G6-G7, G11-G12 and G16-G17 demonstrates that these sequential connectivities are characteristic of 5′-syn-anti-3′ steps in antiparallel topology (Figure 2A).
Three edgewise loops are arranged in an anticlockwise manner forming a ‘-(lll)′ topology (13). Each loop connects the two neighboring G-tracts and consists of 3 nt (1: C3-G4-A5, 2: A8-G9-G10 and 3: C13-G14-T15). A5 and T15 from the first and the third loops are positioned above G2-G6-G12-G16 quartet which is supported by observation of NOE cross-peaks between A5 H8 and G6 H1 as well as between T15 H6 and G16 H1, and T15 Me protons and H1 of G2, G6, G12 and G16 (Figure 2C). The NOE cross-peak between G4 H8 and G6 H8 indicates that G4 residue from the first loop is oriented into the groove. A terminal 3′-tail comprises C18-C19-G20-G21-C22 segment. The NOE cross-peaks between G10 H1 and C18 NH2 suggest formation of G10•C18 base pair in Watson–Crick geometry (Figure 2B). Stacking of G10•C18 base pair below G1-G17-G11-G7 quartet is supported by NOE cross-peaks between G11 H1 and G10 H8, H2′/2’, H5′/5’ as well as between G7 H8 and G10 H8.
A5 is stacked on G2-G6-G12-G16 quartet, while A8 is exposed to solvent
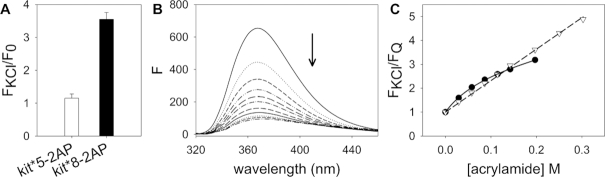
To map the relative positions of adenine residues with respect to G2-G6-G12-G16 and G1-G17-G11-G7 quartets, we alternatively substituted A5 and A8 with a 2-aminopurine (2-AP) in kit*5–2AP and kit*8–2AP without changing the structure (Supplementary Figure S22), respectively. The fluorescence of 2-AP is very sensitive to the environment and, in particular, it is strongly quenched by stacking interactions with guanine residues (44). Fluorescence signals of 2-AP in kit*5–2AP and in kit*8–2AP increase upon addition of 150 mM KCl suggesting that fluorophores are more exposed to solvent in the kit* G-quadruplex in comparison to the pre-folded structure (Figure 3A). Moreover, kit*8–2AP exhibits a significantly higher increase of fluorescence signal compared to kit*5–2AP indicating the tendency of 2-AP in the former oligonucleotide to protrude into solution. To fully investigate this difference, the accessibility of the adenines to solvent was further assessed by monitoring the fluorescence quenching of kit*8–2AP and kit*5–2AP upon titration with acrylamide (Figure 3B). In the case of kit*8–2AP, we observe a linear fluorescence quenching of 2-AP (Figure 3C) and only one Stern–Volmer constant (Ksv) is derived by applying Stern–Volmer formalism (Equation 5). This Ksv value (15.04 ± 0.15 M−1) indicates that A8 experiences only one micro-environment in which it is exposed to solvent and thus, it does not participate in stacking interactions with the G-quartet. On the other hand, kit*5–2AP exhibits a downward curvature of fluorescence quenching plot (Figure 3C) and can be properly fitted using two different Ksv values (Equation 6). The derived value of Ksv1 (1.14 ± 0.51 M−1) corresponds to a micro-environment in which A5 is engaged in stacking interactions with a G-quartet. Conversely, the Ksv2 value (36.06 ± 4.05 M−1) is associated with a position where A5 is more exposed to solvent. Observations of two Ksv values for adenines stacked on a G-quartet, such as A5, have been reported earlier (44). Even though the NMR data indicate a single fold of G-quadruplex, the structural heterogeneity revealed by fluorescence measurements probably results from highly localized, nanosecond fluctuations in positioning of A5 that are confined to the respective loop regions.
Figure 3.
Fluorescence analysis of exposure of 2-AP in kit* to solvent. (A) Fold-change emission of 2-AP in kit*5–2AP and kit*8–2AP at 370 nm upon addition of 150 mM KCl in 10 mM TRIS, pH 7.5 at 25°C. (B) Fluorescence emission variation of 2-AP in kit*8–2AP upon titration with acrylamide in 10 mM TRIS, pH 7.5, 150 mM KCl at 25°C. (C) Fluorescence emission of kit*5–2AP (black dots) and kit*8–2AP (white triangles) at 370 nm plotted as a function of the concentration of acrylamide.
High-resolution structure of kit* G-quadruplex exhibits a well-defined fold-back motif of the 3′-tail
An ensemble of 10 structures of kit* G-quadruplex (Figure 4A and Supplementary Figure S23) was calculated with a simulated-annealing method based on 233 NOE-derived distance, 18 hydrogen bond and 22 torsion-angle restraints (Supplementary Table S2).
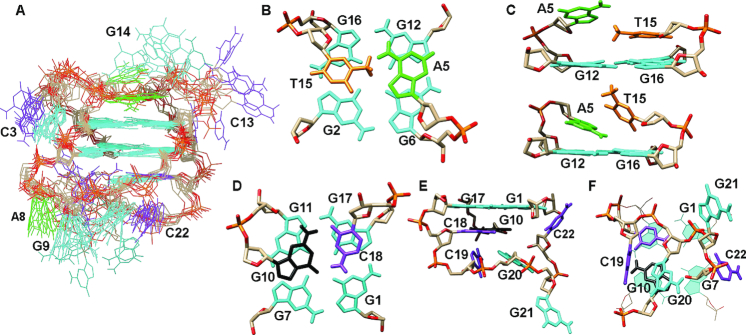
Figure 4.
Structure of kit* G-quadruplex (PDB ID: 6GH0). (A) An ensemble of ten structures. (B and C) Positions of A5 and T15 above G2-G6-G12-G16 quartet. (D) G10•C18 base pair stacked on G1-G17-G11-G7 quartet. (E) Side and (F) bottom view of the fold-back motif of the 3′-tail and stacking of G10•C18 base pair on G1-G17-G11-G7 quartet. Guanines are shown in cyan and black, adenines in green, cytosines in purple and thymines in orange.
The two G-quartet core in the kit* G-quadruplex consisting of G2-G6-G12-G16 (top) and G1-G17-G11-G7 (bottom) quartets is well-defined (RMSD: 0.80 ± 0.32 Å, Figure 4A and Supplementary Figure S23). However, G-quartets are not perfectly planar as G1, G11 and G16 residues are oriented out of the plane for ∼7° (Supplementary Figure S23). A5 and T15 residues adopt three main conformations above the top G-quartet. Importantly, in none of them a base pair is formed between A5 and T15 (Figure 4B and C). In 4 out of 10 structures, A5 and T15 are coplanar and stacked on the top G-quartet. A5 is positioned above G12 and G6 residues, while T15 is stacked over G2 and G16 (Figure 4B). In 6 out of 10 structures only 1 residue, A5 or T15, is stacked on the top G-quartet, while the other is tilted with respect to the G-quartet and exposed to solvent (Figure 4C). Variability in local structure of A5 in kit* G-quadruplex is in good agreement with observation of two Ksv constants in 2-AP fluorescence experiments (vide supra). A5 is part of the first edgewise loop, C3-G4-A5, where G4 is oriented into the groove defined by G2-G6 and G1-G7. C3 is stacked on G4 in 8 out of 10 structures, while it protrudes into solution in the other two. T15 is part of the third edgewise loop, C13-G14-T15, in which G14 is oriented above T15, while C13 protrudes into solution. G14 as well as C13 exhibit much more conformational freedom than T15. On the other side of the G-quartet core, the middle edgewise loop (A8-G9-G10) and the 3′-tail (C18-C19-G20-G21-C22) are oriented below the bottom G-quartet. In particular, G10 and C18 form a well-defined base pair in Watson–Crick geometry, which is stacked on the bottom G-quartet (Figure 4D). Other residues in the 3′-tail, C19-G20-G21-C22, are arranged in a fold-back motif in which C19 is perpendicular to the G10•C18 base pair and G20 is oriented toward it (Figure 4E and F). G21 is oriented in opposite direction compared to the other residues in fold-back motif and protrudes into solution. C22 is positioned perpendicular to the bottom G-quartet in 6 out of 10 structures, while it is oriented into the groove defined by G2-G6 and G1-G7 in 2 out of 10 structures. In other two structures, C22 is oriented away from the bottom G-quartet and stacked on G20 or G21. It is interesting to note that the fold-back motif of the 3′-tail stays preserved also in the course of three independent unrestrained 200 ns MD simulations in the presence of K+ ions and explicit water molecules (Supplementary Figure S24). A8 and G9 residues are part of the middle edgewise loop and are well-defined and oriented toward the residues of the fold-back motif adopted by the 3′-tail (Figure 4). However, both A8 and G9 are exposed to solvent.
Modification and shortening of the 3′-tail crucially alter structure of kit* G-quadruplex, while changes in the loop regions are less significant
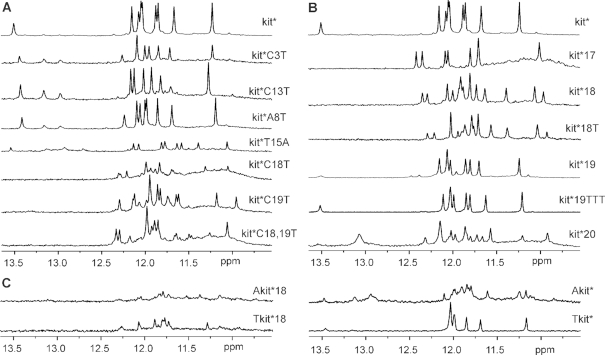
High-resolution structure of kit* G-quadruplex provides detailed information about residues located in the loops and the 3′-tail. To investigate how they influence integrity of kit* G-quadruplex structure we prepared several analogs of kit* with C→T, A→T and T→A modifications in the loops and the 3′-tail (Figure 5A and Supplementary Figure S25). The replacement of C3, A8 and C13 with thymines (i.e. kit*C3T, kit*A8T and kit*C13T) results in eight imino 1H NMR signals characteristic for G-quartet formation and a signal at around δ 13.4 ppm indicating presence of G10•C18 base pair. Similarities between 1H NMR spectra of kit*C3T, kit*A8T and kit*C13T with the parent kit* suggest that these modifications have minimal effect on G-quadruplex structure. For all these modified oligonucleotides we also observe imino signals assigned to pre-kit*. Its imino signals in 1H NMR spectra are more intense in kit*C3T, kit*A8T and kit*C13T compared to kit* indicating the presence of pre-kit* in higher percentage with regards to kit*. Modification T15→A in kit*T15A results in better dispersion of imino signals in comparison to kit*, although G-quadruplex formation is reduced by 70% (Supplementary Figure S26). Modifications of the residues in the 3′-tail in kit*C18T, kit*C19T and kit*C18,19T result in substantial changes in their 1H NMR spectra in comparison to kit*. A hump together with more than eight, broad and overlapped signals in the imino region of 1H NMR spectra of kit*C18T, kit*C19T and kit*C18,19T indicates formation of multiple species, while no imino signal characteristic for G10•C18 base pair is detected. The latter is expected for kit*C18T, but not for kit*C19T, because only C18 is found to be involved in the G10•C18 base pair, stabilizing element of the kit* G-quadruplex.
Figure 5.
Imino region of 1H NMR spectra of (A) modified, (B) 3′-shortened and (C) 5′-extended analogs of kit*. NMR spectra were recorded at ∼0.4 mM concentration of oligonucleotides per strand, 100 mM KCl, pH 7.4 on 800 and 600 MHz NMR spectrometers. Sequences of oligonucleotides are reported in Table 1.
Since sequence modifications in the 3′-tail crucially affect structure of kit* G-quadruplex we prepared six oligonucleotides of different lengths whereupon the 3′-tail has been shortened (Figure 5B, Supplementary Figure S25 and Table 1). 1H NMR spectrum of kit*17 without the entire 3′-tail reveals eight signals between δ 11.00 and 12.49 ppm indicating formation of a G-quadruplex with two G-quartets. No imino signals characteristic of GC base pairs have been observed. At the same time, a hump in the imino region increased within hours indicates slow formation of species with poorly defined structures (vide infra). In the case of kit*18 that contains C18 residue, which is involved in G10•C18 base pair in kit* G-quadruplex, 16 overlapped signals are detected in the imino region of 1H NMR spectrum. This indicates formation of two distinct G-quadruplex structures. It is noteworthy that no signal characteristic for the GC base pair is detected in 1H NMR spectrum of kit*18 at 37°C. Thus, no GC base pair is formed or it is very exposed to solvent because it is not protected by the 3′-tail. In kit*18T an additional thymine residue at the 3′-end does not induce formation of expected GC base pair or lead to a single G-quadruplex structure. In the case of kit*19, eight imino signals and a broad signal at δ 13.51 ppm were observed in 1H NMR spectrum. Moreover, we detected at least four additional imino signals with lower intensity indicating the presence of a minor G-quadruplex structure in the sample of kit*19. Formal extension with three additional thymine residues at the 3′-end was favorable for the formation of one G-quadruplex structure with G10•C18 base pair as revealed by nine (8 + 1) imino signals in 1H NMR spectrum of kit*19TTT. 3′-TTT end helps to stabilize kit* G-quadruplex structure through additional interactions with residues from the second loop similarly as G20-G21-C22 fragment in kit*. Formation of multiple G-quadruplex species was observed for kit*20.
Extension of oligonucleotide kit*18 at the 5′-end with thymine (i.e. Tkit*18) or adenine (i.e. Akit*18) residues reduced formation of G-quadruplexes by 80% as revealed by weak signals in the imino region of 1H NMR spectrum (Figure 5C, Supplementary Figure S25 and Table 1). Interestingly, for Tkit*18 eight imino signals detected in 1H NMR spectrum suggest presence of one major G-quadruplex. On the contrary, extension at the 5′-end of kit* with thymine (i.e. Tkit*) was not critical for G-quadruplex structure. Elongation with adenine residue (i.e. Akit*) of kit* is more relevant in the natural context of c-KIT gene, but weak imino signals in 1H NMR spectrum indicating that is less favorable for the formation of G-quadruplex. We detected eight resolved and many broad imino signals between δ 10.9 and 12.1 ppm in 1H NMR spectrum of Akit* suggesting formation of two G-quadruplex structures. Signal at δ 13.48 ppm assigned to G10•C18 base pair was also detected in addition to signals of pre-folded structure at around δ 13.0 ppm.
The folding pathway is affected by the length of kit*
To better unveil the role of the 3′-tail in the G-quadruplex folding pathway we performed CD kinetic experiments on kit*, kit*17, kit*18 and kit*19. Upon the addition of 150 mM KCl, kit*, kit*18 and kit*19 exhibit fast folding kinetic into antiparallel G-quadruplexes that is completed within the sample mixing time of 10 s (Supplementary Figure S27A–C). Although NMR data show that kit*18 and kit*19 fold into two distinct G-quadruplex structures each, the CD spectra of equilibrated kit*, kit*18 and kit*19 overlap. This suggests that all their G-quadruplexes share a common antiparallel topology.
The behavior of kit*17 is different. This oligonucleotide initially folds into an antiparallel G-quadruplex (Supplementary Figure S27D, gray solid line), which then slowly evolves to other folded species, as revealed by CD signal variations over time (Supplementary Figure S27D, black dashed line). The folding pathway of kit*17 was studied by applying singular value decomposition analysis (45,46) to the CD dataset of its folding process (Supplementary Figures S27D and 28). The initial folding stage (F0→F1) is a fast event, thus its accurate analysis was not possible with our equipment (Supplementary Figure S28D). For this reason, we analyzed only the slower folding step (F1→F2) (Supplementary Figure S28D). Singular values in the S matrix and the U and V autocorrelation coefficients (meaningful above 0.75) indicate that, in the F1→F2 folding process, two main species contribute to the dichroic signal variation (Supplementary Figure S28A). Significant V eigenvectors were fitted by applying a mono-exponential kinetic model (Equation 2, Supplementary Figure S28B). The fitting parameters allowed us to obtain the basic spectra of the species in solution (Supplementary Figure S28C). F1 displays the dichroic spectrum typical of an antiparallel G-quadruplex (Supplementary Figure S28C, solid line). It is formed immediately upon addition of KCl and it slowly converts into F2, whose dichroic spectrum presents a positive peak at 264 nm and a negative one at 248 nm consistent with a parallel G-quadruplex arrangement (Supplementary Figure S28C, dashed line). The time needed to achieve the equilibrium (τ) of the F1→F2 process is 4.0 ± 0.2 h.
CD melting experiments reveal that kit* exhibits a clear single-transition melting profile (Figure 6) with a melting temperature (Tm) of 60.5 ± 0.1°C in 150 mM KCl (Equation 3). This observations suggest that kit* folds mainly into one conformation at these experimental conditions. In comparison, kit*17 melting profile displays two different melting transitions (Figure 6 and Equation 4) at 50.6 ± 0.6°C (Tm1) and at 75.9 ± 0.4°C (Tm2) confirming that at least two structures are present in the solution and participate in the melting process. Tm1 is associated with the kinetically favored antiparallel G-quadruplex, while Tm2 is related to thermodynamically favored parallel forms. Interestingly, melting of antiparallel kit*17 G-quadruplex can be followed through the increase of CD melting profile, because its negative optical contribution in CD spectra at 264 nm is reduced (Figure 6C). At 56°C more stable parallel structure starts to melt, which is reflected in decreasing CD melting profile at 264 nm. We propose that Tm1 of kit*17 is lower compared to the Tm of the kit* structure because of the lack of the 3′-tail which contributes to the stability of the entire G-quadruplex. UV melting analysis acquired on kit*18 and kit*19 at experimental conditions used for NMR experiments revealed comparable results as CD melting data. Two thermal transitions are detected also for kit*18, while kit*19 shows a reduction of the thermal stability by 3°C compared to kit* (Supplementary Figure S29). These data fully confirm the destabilizing influence of shortening of the 3′-tail in kit*.
Figure 6.
Temperature-dependent variation between 25 (black solid line) and 95°C (black-dashed line) of CD spectra of (A) kit* and (B) kit*17 in 150 mM KCl. (C) Melting profile of kit* (black dots) and kit*17 (white triangles) monitored at 294 and 264 nm, respectively.
Two molecules of kit* are partially paired in a homo-dimeric structure of pre-folded structure
Two signals at δ 12.94 and 13.14 ppm in 1H NMR spectrum at 37°C of kit* in the presence of Li+ ions indicate formation of a pre-folded structure, named pre-kit*, that does not contain stacked G-quartets (Figure 7A). Unambiguous assignment of imino signals, using residue-specific, partially 13C,15N-labeled kit*, reveals that G20 and G21 residues of the 3′-tail are involved in GC Watson–Crick base pairs in pre-kit* (Supplementary Figure S30). The base pairing in pre-kit* occurs between G20 and G21 of one molecule and C18 and C19 of another kit* molecule (Supplementary Figure S31). Thus, the proposed partially paired segment of kit* molecules consists of four GC base pairs and is highly symmetric, which is in agreement with the observation of only two imino signals in 1H NMR spectrum. CD spectrum of pre-kit* in the presence of Li+ ions at 25°C displays a maximum at 284 nm and a minimum at 245 nm confirming an antiparallel double-stranded secondary structure (Figure 1C and Supplementary Figure S32). Existence of an isodichroic point at 255 nm suggests that there is only one conformational step from unfolded oligonucleotide toward pre-kit* form. Noteworthy, decrease of temperature of the Li+ ion containing solution from 37 to 5°C enables detection of pre-folded structures, besides pre-kit*, that are involved in conformational exchange. Indeed, at 37°C only signals in 1H NMR spectrum corresponding to pre-kit* are observed, while upon lowering the sample temperature to 5°C new signals appear between δ 12.2–13.8 and 10.2–11.8 ppm in 1H NMR spectrum (Figure 7B). In 15N-edited HSQC NMR spectra in the presence of LiCl at 5°C two imino signals were observed for every residue of kit* that was 13C,15N-labeled (Supplementary Figure S33). These results suggest coexistence of two different secondary structures in the presence of Li+ ions at 5°C. Pre-kit* is mostly stabilized by GC base pairs, while the other structure contains more base pairs in non–Watson–Crick geometry.
Figure 7.
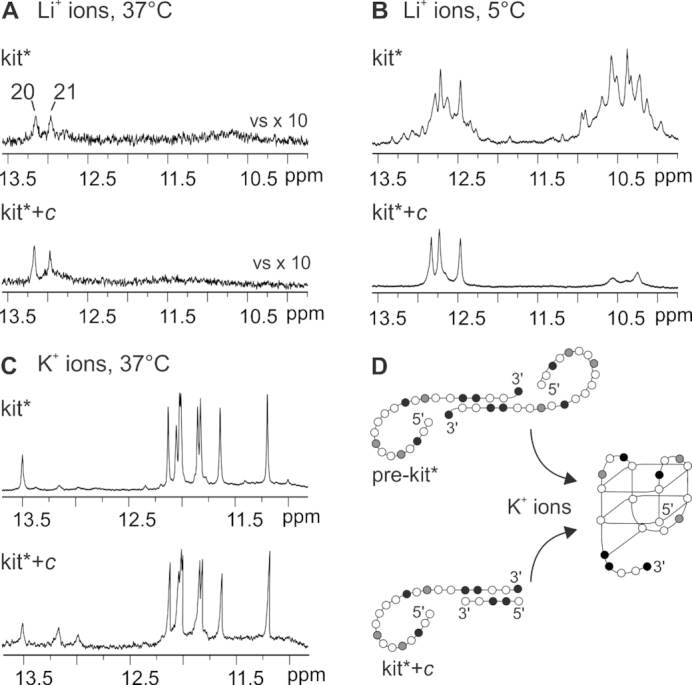
Imino region of 1H NMR spectra of kit* and equimolar mixture of kit* and construct c (kit*+c) recorded in the presence of Li+ ions at (A) 37 and (B) 5°C as well as (C) in the presence of K+ ions at 37°C. (D) Schematic presentation of forms involved in formation of kit* G-quadruplex. Guanines are marked in white, cytosines in black, adenines and thymines in gray.
In order to explore interactions of residues in the 3′-tail of pre-kit* structure, we designed a 5 nt long construct c, d[GCCGG], that is complementary to C18-C19-G20-G21-C22 segment of kit*. Our results show that base pairing of the 3′-tail of kit* with the construct c blocks its other possible interactions (Figure 7B). 1H NMR spectrum of kit* hybridized with the construct c (marked as kit*+c) in equimolar amounts displays similar pattern of imino signals compared to kit* at 37°C (Figure 7A). Upon lowering the temperature to 5°C, three well-resolved imino signals between δ 12.4 and 13.0 ppm have been detected for kit*+c instead of many overlapped peaks in the presence of Li+ ions for kit* (Figure 7B). Formation of self-complementary structures adopted by construct c was not observed (Supplementary Figure S34). Interestingly, in the case of kit* as well as kit*+c formation of the same G-quadruplex is detected after addition of K+ ions (Figure 7C). These results indicate that kit* in the pre-folded structure pre-kit* and kit*+c share a comparable inter-molecular arrangement of the 3′-tail in the presence of LiCl, where the 3′-tails of two distinct kit* molecules are paired through GC Watson–Crick interactions forming a homo-dimer (Figure 7D and Supplementary Figure S5). Signals assigned to pre-folded structures were observed in NMR spectra of kit* as well as kit*+c also in the presence of K+ ions (Figure 7C and Supplementary Figure S35).
DISCUSSION
c-KIT proto-oncogene exhibits three G-rich regions in its promoter, kit1, kit* and kit2, that are able to fold into G-quadruplexes. kit* region plays a key role in regulation of c-KIT transcription since it contains consensus sites for SP1 and AP2 transcription factors. The NMR derived kit* structure presented herein is, to the best of our knowledge, the first high-resolution structure of a two G-quartet G-quadruplex originating from a promoter region. At physiologically relevant temperature (37°C) and concentration of K+ ions (100 mM) kit* adopts a chair type antiparallel G-quadruplex comprising three edgewise loops and a 3′-tail. A5 and T15 residues from the first and the third loops are co-planar and stacked on the top G-quartet, or only one of them is stacked on the top G-quartet, while alternatively the other residue protrudes into solution. Stacking of adenine and thymine from two edgewise loops on outer G-quartet was observed before (47), where A and T face each other with their Watson–Crick edges. This is very different relative orientation above the outer G-quartet compared to A5 and T15 in kit* G-quadruplex. It is interesting, that in both cases AT base pairs were not formed. Other residues from the first and the third loops of kit* protrude into solution or are oriented into the grooves. On the other side of the kit* G-quartet core, G10•C18 base pair is stacked on the bottom G-quartet. C19, G20, G21 and C22 residues from the 3′-tail are arranged into a fold-back motif below the G10•C18 base pair and the bottom G-quartet. To compare, in the case of two G-quartet G-quadruplexes originating from telomeric regions base triples are stacked on both G-quartets (48–51). The fold-back motif consisting of three terminal residues, in which two of them are stacked on G-quartet, has been described for G-quadruplex originating from human c-MYC promoter (52). Four residue overhang has been shown to adopt an extra-G-quartet fold-back element that contributes to stabilization of VEGF aptamer (53).
kit* in its sequence contains one GGGG-tract and four GG-repeats, of which G20-G21 repeat, located in the 3′-terminal, is not involved in the formation of G-quartet core. Contrary to expectations, we observed that 3′-tail plays an important role in the folding process, structural integrity and thermal stability of kit* G-quadruplex. First, the 3′-tail is involved in the formation of a stable pre-folded structure in the absence of K+ ions. In this form, residues in the 3′-tail of two kit* molecules are base paired through GC Watson–Crick interactions forming an inter-molecular partially paired homo-dimeric structure. Upon addition of K+ ions, organization of the 3′-tail in pre-kit* prevents Hoogsteen base pairing of the terminal GG-repeat (G20-G21). It is noteworthy that modifications in the 3′-tail disturb GC base pairs in pre-kit* and as a result lead to formation of more than two G-quadruplexes. Once G1-G17 residues of kit* get involved in the G-quadruplex, the preservation of GC base pairing of two 3′-tails is most likely sterically unfavored and the 3′-tails dissociate from the complementary strand leading to formation of a monomeric G-quadruplex. In full agreement, the replacement of G20-G21 repeat with thymine residues results in one G-quadruplex structure. Second, G10•C18 base pair and the fold-back motif of the 3′-tail stabilize kit* G-quadruplex by covering the bottom G-quartet. Perusal of the high-resolution structure of kit* G-quadruplex show that the fold-back structural element enables additional hydrogen bonds, electrostatic and hydrophobic interactions between residues of the 3′-tail and the second loop as well as the bottom G-quartet. Examination of the folding behavior has revealed that the 3′-tail plays an important role in folding of kit*. For kit*17 without the 3′-tail alteration of folding pathway was observed together with formation of antiparallel structure and poorly defined G-quadruplexes with parallel topology that were formed within few hours after addition of K+ ions. It is noteworthy that even preservation of the G10•C18 stabilizing element alone is not sufficient for the formation of a single kit* G-quadruplex. Thus, the conformational selection of only one kit* G-quadruplex is controlled by the presence of all residues of the 3′-tail in kit*. These observations are unique as terminal regions often change folding kinetics of oligonucleotide and destabilize formation of the final G-quadruplex structure (54–56). The elongation of kit* with thymine residue at the 5′-end also did not disturb kit* G-quadruplex structure, while adenine residue is less favorable and lead to formation of additional minor structure.
Considering the whole G-rich region in promoter of c-KIT gene, kit1 and kit2 G-rich oligonucleotides in contrast to kit* fold into three G-quartet parallel G-quadruplexes. It is worth mentioning that differences in structures of kit1, kit2 and kit* G-quadruplexes are associated with differences of kinetics of their formation. kit* folds in seconds, which is on a timescale of transcriptional processes (54,57). This is faster compared to folding kinetics of kit2, where the initial fast folding kinetic intermediate converts into the thermodynamically stable structures only within hours (34). Kinetic data for kit1 are not available, but since it forms a parallel G-quadruplex slower folding kinetic can be predicted compared to antiparallel kit* G-quadruplex (54). Therefore, kit* could modulate the conformational organization of the neighboring kit1 and kit2 regions and thus plays a functional role in regulation of transcription. Structural features of G-rich regions in promoter of c-KIT gene have been recognized for their interactions with zinc finger motifs of proteins such as SP1 (7,58–60). The dynamic interplay of c-KIT gene promoter suggests that kit* could serve as an optimal site of intervention to finely tune the level of c-kit expression. Interestingly, this potential regulatory role merges with the complexity of the herein solved structure where the stability of the kit* G-quadruplex depends upon a network of interactions involving the loops and the 3′-tail. The structural features allow for foreseeing a possibility of selective recognition where ligands with even modest binding affinity may be able to induce cellular effects. Notably, selective stabilization of c-KIT compared to human telomeric G-quadruplex has been already reported for isoalloxazines ligands (61). Discrimination of ligands between kit1 and kit2 G-quadruplexes has been reported showing specific recognition and structural stabilization (61–64). Structural data of kit* presented herein provides new insights for design of ligands that can target the G-rich region of c-KIT gene more comprehensively and with higher specificity, which could result in reduction of frequently occurring off-targeting of multiple G-quadruplexes (65).
In conclusion, structural characterization of the kit* G-quadruplex, the identification of a pre-folded structure and insights into its folding behavior expand on our knowledge about the processes involved in the regulation of the c-KIT proto-oncogene transcription. In particular, faster folding kinetic of kit* in comparison to kit1 and kit2 G-quadruplexes raises new questions about the role of kit* in affecting the folding processes of neighboring G-rich regions. At last, the high-resolution structure of kit* G-quadruplex with two G-quartet antiparallel topology, the GC base pair and the fold-back motif of the 3′-tail, can be a suitable starting point for rational design of tailor-made ligands that will modulate c-kit expression.
DATA AVAILABILITY
The coordinates for structure of kit* G-quadruplex have been deposited in the Protein Data Bank (accession number: 6GH0) and Biological Magnetic Resonance Data Bank (accession number: 34269).
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the CERIC-ERIC Consortium for the access to experimental facilities and financial support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Slovenian Research Agency [P1-0242, J1-7108 to A.K., J.P.]; CERIC-ERIC; University of Padova [CPDA147272/14 to C.S.]; University of Padova Ph.D. Fellowship (to R.R.). Funding for open access charge: Slovenian Research Agency [P1-0242].
Conflict of interest statement. None declared.
REFERENCES
- 1. d’Auriol L., Mattei M.G., Andre C., Galibert F.. Localization of the human c-kit protooncogene on the q11-q12 region of chromosome 4. Hum. Genet. 1988; 78:374–376. [DOI] [PubMed] [Google Scholar]
- 2. Yarden Y., Kuang W.J., Yang-Feng T., Coussens L., Munemitsu S., Dull T.J., Chen E., Schlessinger J., Francke U., Ullrich A.. Human proto-oncogene c-kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987; 6:3341–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yamamoto K., Tojo A., Aoki N., Shibuya M.. Characterization of the promoter region of the human c-kit proto-oncogene. Jpn. J. Cancer Res. 1993; 84:1136–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Metcalfe D.D. Mast cells and mastocytosis. Blood. 2008; 112:946–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gregory-Bryson E., Bartlett E., Kiupel M., Hayes S., Yuzbasiyan-Gurkan V.. Canine and human gastrointestinal stromal tumors display similar mutations in c-KIT exon 11. BMC Cancer. 2010; 10:559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rankin S., Reszka A.P., Huppert J., Zloh M., Parkinson G.N., Todd A.K., Ladame S., Balasubramanian S., Neidle S.. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005; 127:10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raiber E.A., Kranaster R., Lam E., Nikan M., Balasubramanian S.. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012; 40:1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hud N.V., Plavec J.. The Role of Cations in Determining Quadruplex Structure and Stability in Quadruplex Nucleic Acids. 2006; Cambridge: The Royal Society of Chemistry. [Google Scholar]
- 9. Fujii T., Podbevšek P., Plavec J., Sugimoto N.. Effects of metal ions and cosolutes on G-quadruplex topology. J. Inorg. Biochem. 2017; 166:190–198. [DOI] [PubMed] [Google Scholar]
- 10. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karsisiotis A.I., O’Kane C., Webba da Silva M.. DNA quadruplex folding formalism—a tutorial on quadruplex topologies. Methods. 2013; 64:28–35. [DOI] [PubMed] [Google Scholar]
- 12. Šket P., Plavec J.. Tetramolecular DNA quadruplexes in Solution: insights into structural diversity and cation movement. J. Am. Chem. Soc. 2010; 132:12724–12732. [DOI] [PubMed] [Google Scholar]
- 13. Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chem. Eur. J. 2007; 13:9738–9745. [DOI] [PubMed] [Google Scholar]
- 14. Greco M.L., Kotar A., Rigo R., Cristofari C., Plavec J., Sissi C.. Coexistence of two main folded G-quadruplexes within a single G-rich domain in the EGFR promoter. Nucleic Acids Res. 2017; 45:10132–10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marušič M., Šket P., Bauer L., Viglasky V., Plavec J.. Solution-state structure of an intramolecular G-quadruplex with propeller, diagonal and edgewise loops. Nucleic Acids Res. 2012; 40:6946–6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shivalingam A., Izquierdo M.A., Marois A.L., Vysniauskas A., Suhling K., Kuimova M.K., Vilar R.. The interactions between a small molecule and G-quadruplexes are visualized by fluorescence lifetime imaging microscopy. Nat. Commun. 2015; 6:8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M. Jr, Sen D., Lansdorp P.M.. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014; 42:860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hansel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017; 18:279–284. [DOI] [PubMed] [Google Scholar]
- 20. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotech. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 21. Maizels N., Gray L.T.. The G4 genome. PLoS Genet. 2013; 9:e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huppert J.L., Balasubramanian S.. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007; 35:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017; 1:41. [Google Scholar]
- 24. Rigo R., Palumbo M., Sissi C.. G-quadruplexes in human promoters: a challenge for therapeutic applications. Biochim. Biophys. Acta. 2017; 1861:1399–1413. [DOI] [PubMed] [Google Scholar]
- 25. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collie G.W., Parkinson G.N.. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011; 40:5867–5892. [DOI] [PubMed] [Google Scholar]
- 27. Xu H., Di Antonio M., McKinney S., Mathew V., Ho B., O’Neil N.J., Santos N.D., Silvester J., Wei V., Garcia J. et al.. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017; 8:14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gunaratnam M., Swank S., Haider S.M., Galesa K., Reszka A.P., Beltran M., Cuenca F., Fletcher J.A., Neidle S.. Targeting human gastrointestinal stromal tumor cells with a quadruplex-binding small molecule. J. Med. Chem. 2009; 52:3774–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McLuckie K.I., Waller Z.A., Sanders D.A., Alves D., Rodriguez R., Dash J., McKenzie G.J., Venkitaraman A.R., Balasubramanian S.. G-quadruplex-binding benzo[a]phenoxazines down-regulate c-KIT expression in human gastric carcinoma cells. J. Am. Chem. Soc. 2011; 133:2658–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zorzan E., Da Ros S., Musetti C., Shahidian L.Z., Coelho N.F., Bonsembiante F., Letard S., Gelain M.E., Palumbo M., Dubreuil P. et al.. Screening of candidate G-quadruplex ligands for the human c-KIT promotorial region and their effects in multiple in-vitro models. Oncotarget. 2016; 7:21658–21675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hsu S.T., Varnai P., Bugaut A., Reszka A.P., Neidle S., Balasubramanian S.. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 2009; 131:13399–13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Phan A.T., Kuryavyi V., Burge S., Neidle S., Patel D.J.. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007; 129:4386–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuryavyi V., Phan A.T., Patel D.J.. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010; 38:6757–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rigo R., Dean W.L., Gray R.D., Chaires J.B., Sissi C.. Conformational profiling of a G-rich sequence within the c-KIT promoter. Nucleic Acids Res. 2017; 45:13056–13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller M.C., Le H.T., Dean W.L., Holt P.A., Chaires J.B., Trent J.O.. Polymorphism and resolution of oncogene promoter quadruplex-forming sequences. Org. Biomol. Chem. 2011; 9:7633–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Da Ros S., Zorzan E., Giantin M., Zorro Shahidian L., Palumbo M., Dacasto M., Sissi C.. Sequencing and G-quadruplex folding of the canine proto-oncogene KIT promoter region: might dog be used as a model for human disease. PLoS One. 2014; 9:e103876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wei D., Husby J., Neidle S.. Flexibility and structural conservation in a c-KIT G-quadruplex. Nucleic Acids Res. 2015; 43:629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wei D., Parkinson G.N., Reszka A.P., Neidle S.. Crystal structure of a c-kit promoter quadruplex reveals the structural role of metal ions and water molecules in maintaining loop conformation. Nucleic Acids Res. 2012; 40:4691–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park G.H., Plummer H.K., Krystal G.W.. Selective Sp1 binding is critical for maximal activity of the human c-kit promoter. Blood. 1998; 92:4138–4149. [PubMed] [Google Scholar]
- 40. Rigo R., Sissi C.. Characterization of G4-G4 crosstalk in the c-KIT promoter region. Biochemistry. 2017; 56:4309–4312. [DOI] [PubMed] [Google Scholar]
- 41. Kocman V., Plavec J.. Tetrahelical structural family adopted by AGCGA-rich regulatory DNA regions. Nat. Commun. 2017; 8:15355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kotar A., Wang B., Shivalingam A., Gonzalez‐Garcia J., Vilar R., Plavec J.. NMR structure of a triangulenium‐based long‐lived fluorescence probe bound to a G‐quadruplex. Angew. Chem. Int. Ed. Engl. 2016; 55:12508–12511. [DOI] [PubMed] [Google Scholar]
- 43. Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A.. Development and testing of a general amber force field. J. Comput. Chem. 2004; 25:1157–1174. [DOI] [PubMed] [Google Scholar]
- 44. Gray R.D., Petraccone L., Buscaglia R., Chaires J.B.. 2-aminopurine as a probe for quadruplex loop structures. Methods Mol. Biol. 2010; 608:121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hendler R.W., Shrager R.I.. Deconvolutions based on singular value decomposition and the pseudoinverse: a guide for beginners. J. Biochem. Biophys. Methods. 1994; 28:1–33. [DOI] [PubMed] [Google Scholar]
- 46. DeSa R.J., Matheson I.B.. A practical approach to interpretation of singular value decomposition results. Methods Enzymol. 2004; 384:1–8. [DOI] [PubMed] [Google Scholar]
- 47. Wang Y., Patel D.J.. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993; 1:263–282. [DOI] [PubMed] [Google Scholar]
- 48. Galer P., Wang B., Sket P., Plavec J.. Reversible pH switch of two-quartet G-quadruplexes formed by human telomere. Angew. Chem. Int. Ed. Engl. 2016; 55:1993–1997. [DOI] [PubMed] [Google Scholar]
- 49. Lim K.W., Amrane S., Bouaziz S., Xu W., Mu Y., Patel D.J., Luu K.N., Phan A.T.. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009; 131:4301–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Z., Dai J., Veliath E., Jones R.A., Yang D.. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010; 38:1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Amrane S., Ang R.W., Tan Z.M., Li C., Lim J.K., Lim J.M., Lim K.W., Phan A.T.. A novel chair-type G-quadruplex formed by a Bombyx mori telomeric sequence. Nucleic Acids Res. 2009; 37:931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ambrus A., Chen D., Dai J., Jones R.A., Yang D.. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005; 44:2048–2058. [DOI] [PubMed] [Google Scholar]
- 53. Marušič M., Veedu R.N., Wengel J., Plavec J.. G-rich VEGF aptamer with locked and unlocked nucleic acid modifications exhibits a unique G-quadruplex fold. Nucleic Acids Res. 2013; 41:9524–9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Marchand A., Gabelica V.. Folding and misfolding pathways of G-quadruplex DNA. Nucleic Acids Res. 2016; 44:10999–11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gray R.D., Chaires J.B.. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008; 36:4191–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Arora A., Nair D.R., Maiti S.. Effect of flanking bases on quadruplex stability and Watson-Crick duplex competition. FEBS J. 2009; 276:3628–3640. [DOI] [PubMed] [Google Scholar]
- 57. Morisaki T., Muller W.G., Golob N., Mazza D., McNally J.G.. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat. Commun. 2014; 5:4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Elrod-Erickson M., Rould M.A., Nekludova L., Pabo C.O.. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996; 4:1171–1180. [DOI] [PubMed] [Google Scholar]
- 59. Narayan V.A., Kriwacki R.W., Caradonna J.P.. Structures of zinc finger domains from transcription factor Sp1. Insights into sequence-specific protein-DNA recognition. J. Biol. Chem. 1997; 272:7801–7809. [DOI] [PubMed] [Google Scholar]
- 60. Oka S., Shiraishi Y., Yoshida T., Ohkubo T., Sugiura Y., Kobayashi Y.. NMR structure of transcription factor Sp1 DNA binding domain. Biochemistry. 2004; 43:16027–16035. [DOI] [PubMed] [Google Scholar]
- 61. Bejugam M., Sewitz S., Shirude P.S., Rodriguez R., Shahid R., Balasubramanian S.. Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression. J. Am. Chem. Soc. 2007; 129:12926–12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Duarte A.R., Cadoni E., Ressurreicao A.S., Moreira R., Paulo A.. Design of modular G-quadruplex ligands. Chemmedchem. 2018; 13:869–893. [DOI] [PubMed] [Google Scholar]
- 63. Diveshkumar K.V., Sakrikar S., Rosu F., Harikrishna S., Gabelica V., Pradeepkumar P.I.. Specific stabilization of c-MYC and c-KIT G-quadruplex DNA structures by indolylmethyleneindanone scaffolds. Biochemistry. 2016; 55:3571–3585. [DOI] [PubMed] [Google Scholar]
- 64. Bejugam M., Gunaratnam M., Muller S., Sanders D.A., Sewitz S., Fletcher J.A., Neidle S., Balasubramanian S.. Targeting the c-Kit promoter G-quadruplexes with 6-Substituted indenoisoquinolines. ACS Med. Chem. Lett. 2010; 1:306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vilar R. Nucleic acid quadruplexes and Metallo-Drugs. Met. Ions Life Sci. 2018; 18:325–349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates for structure of kit* G-quadruplex have been deposited in the Protein Data Bank (accession number: 6GH0) and Biological Magnetic Resonance Data Bank (accession number: 34269).