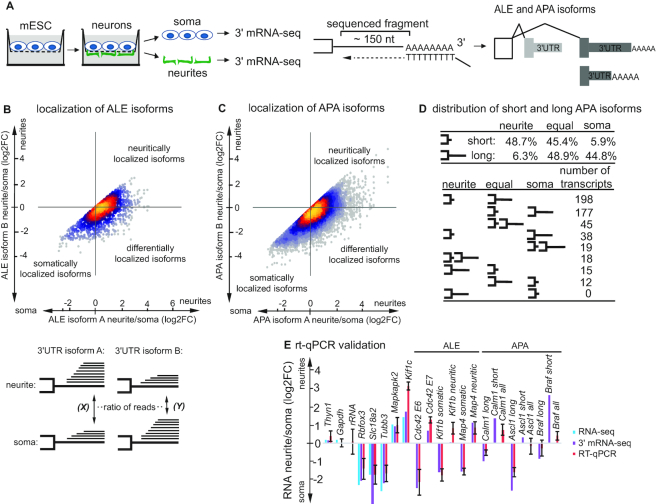
Figure 1.
Alternative polyadenylation sites usage and resulting alternative 3′UTRs affect mRNA localization to neurites. (A) Scheme of neurite/soma separation and 3′ mRNA-seq. mESC-derived neurons are grown on a microporous membrane so that neurites extend on the lower side of the membrane to enable separation of the soma and neurites. RNA isolated from subcellular compartments is subjected to 3′ mRNA-seq, which is based on oligodT priming and enables amplification and sequencing of polyadenylated mRNA 3′ ends. The resulting data are analyzed for the ALE and APA isoforms. (B) Scatterplot illustrating differential localization patterns for transcripts with ALE 3′UTR isoforms. 3′UTR isoforms are designated as A and B on the condition that A is equally or more represented in neurites than B (see the scheme below the plot). Enrichment in neurites for isoform A (X) is plotted against the same enrichment for isoform B (Y). Dots falling on diagonal correspond to genes with similar localization patterns of alternative 3′UTR isoforms. For the rest of the genes, transcripts with alternative 3′UTRs show differential localization between neurites and soma. Coloring indicates local point density. (C) Plot illustrating differential localization patterns for transcripts with APA 3′UTR isoforms. Data are presented as in (B). (D) Distribution of short and long APA isoforms between neurites and soma. The top panel shows percentages of short (]-) and long (]—) APA isoforms, which are enriched in neurites, enriched in soma, or else equally distributed. The lower panel shows localization patterns of short and long APA isoforms deriving from the same gene. Neurites: enriched in neurites >2-fold; equal: <2-fold change between neurites and soma; soma: enriched in soma >2-fold. (E) qRT-PCR for selected differentially localized alternative 3′UTRs. Error bars represent SD for two (neurites) to three (soma) biological replicates. Gapdh (reference RNA), Thyn1, rRNA were used as unlocalized controls, Rbfox3, Slc18a2, Tubb3 as soma-localized, Mapkap2, Kif1c as neurite-localized controls. ENSEMBL identifiers of the isoforms: Cdc42E6 ENSMUST00000030417.9; Cdc42E7 ENSMUST00000051477.12; Kif1b somatic ENSMUST00000060537.12; Kif1b neuritic ENSMUST00000030806.5; Map4 somatic ENSMUST00000169851.7; Map4 neuritic ENSMUST00000035055.14. For a reference, we also show neurite/soma enrichment based on 3′ mRNA-seq (purple bars) and RNA-seq data (blue bars, not isoform-specific) next to the qRT-PCR data (red bars). In case of APA isoforms, the qRT-PCR data are shown for a long isoform and both isoforms combined (all), as short APA isoforms cannot be distinguished from long APA isoforms by qPCR.