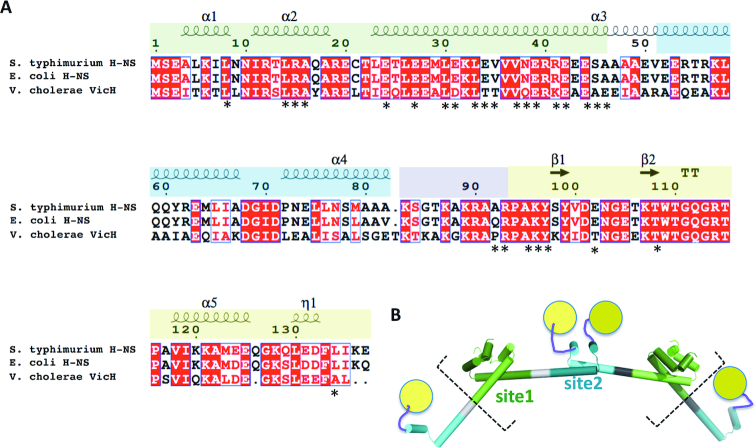
Figure 1.
Domain organization of H-NS. (A) Sequence alignment showing structural elements (α-helices, β-strands; η represents a 310 helix). Protein domains (determined from experimental structures) are green: N-terminal dimerization domain (site1); blue: central dimerization domain (site2), purple: positively charged linker; yellow: C-terminal DNA binding domain (DNAbd). Positions of S. typhimurium residues showing chemical shift changes in presence of H-NS84–137 or H-NS2–57 are indicated by an asterisk. (B) Schematic structural model for multimerization through site1 (light and dark green) and site2 (light and dark cyan) extracted from the H-NS superhelix conformation (see (35) and PDB 3NR7). The charged linker (purple) and DNAbd (yellow) are not included in the crystallographic model. Dashed square brackets outline the construct used in MD simulations.