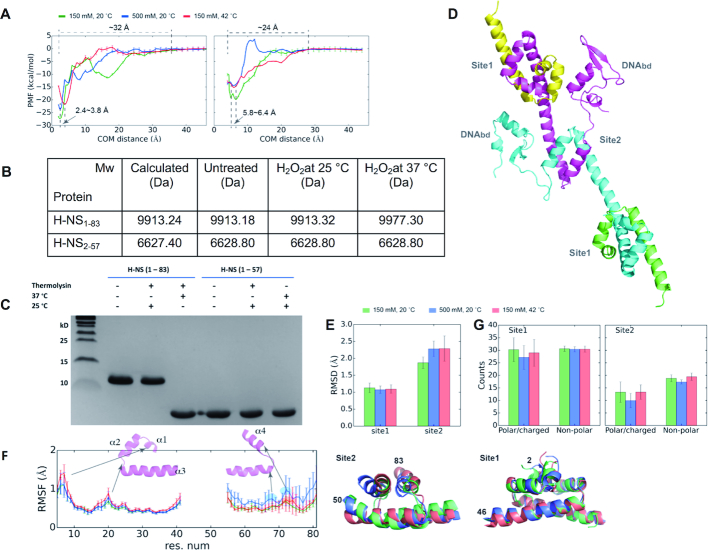
Figure 3.
Site2, but not site1, of S. typhimurium H-NS is sensitive to changes in temperature and salinity. (A) PMF plots of site1 (left) and site2 (right) dimers along the pulling direction using COM distance between two monomers as the collective variable. The dissociation state of the monomers is set as the reference (0.0 kcal/mol) for each calculation. We averaged the data with SDs as error bars from six PMFs; the small standard deviations (SDs) indicate the convergence of the PMF. (B) Molecular weight determination of H-NS1–83,C21S or H-NS2–57,C21S at different temperatures in the presence or absence of hydrogen peroxide (H2O2). Calculated Mw for unmodified proteins includes leftover of cleavage from tags (H-NS1–83,C21S: residues GPLGS; H-NS2–57: GS). (C) Proteolytic cleavage of H-NS1–83 or H-NS2–57 with thermolysin at different temperatures. (D) A final snapshot of the full-length H-NS dimer (centre), and superimposed site1 (top) and site 2 (bottom) structures under regular, salt, and heat conditions taken from 200 ns MD simulations. (E) Histogram of Cα RMSD of site1 and site2 dimers, with the S. typhimurium H-NS crystal structure (PDBID: 3NR7) as the reference. We superposed residues 5–41 and residues 55–81 for the RMSD calculations of site1 and site2, respectively. (F) Plot of average residue fluctuation (RMSF) for site1 and site2 with respect to the crystal structure (PDBID: 3NR7), presented along the H-NS sequence. Prominent features the high-temperature and high-salinity conditions are highlighted in red and blue, respectively. (G) Histograms of the polar(H-bond)/charged(ionic) contacts and of non-polar(hydrophobic) contacts across protein chains of site1 and site2 dimers. The data in (E) and (G) were averaged with SDs as error bars from all replica simulations.