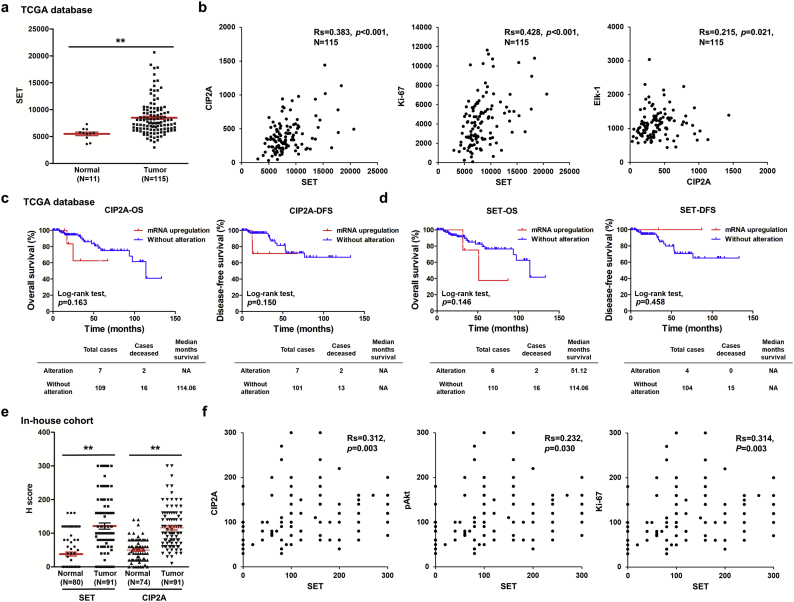
Fig. 1.
SET and CIP2A were up-regulated in the TNBC tumor tissues and correlated with CIP2A and Ki-67. (a-b) Level 3 data of mRNA expressions from TNBC samples and normal tissue samples were downloaded from the TCGA and Broad GDAC Firehose data portal. The mRNA RSEM of all samples were selected and analyzed for comparing abundances by GraphPad Prism 5 software. The transcript levels of SET, CIP2A, Ki-67 and Elk-1 were measured by RNA sequencing in TCGA data. The transcript levels of SET in TNBC tumor samples and normal tissue samples were measured by RNA sequencing in TCGA data (a). The correlation of SET, CIP2A, Ki-67 and Elk-1 were analyzed. Rs, Spearman's rank correlation coefficient (b). (c-d) Overall survival (OS) and disease-free survival (DFS) curves were plotted for TNBC cases with or without alteration of SET and CIP2A expressions. (e-f) The tissue sections were quantitatively scored according to the percentage of positive cells and staining intensity. SET and CIP2A expression scores in TNBC tissue specimens and normal tissues (e). The relative protein levels of CIP2A, pAkt and Ki-67 in TNBC tumor samples were positively correlated with those of SET levels (f). Rs, Spearman's rank correlation coefficient. Student's t-test, *, P < 0.05; **, P < 0.01; ***, P < 0.001.