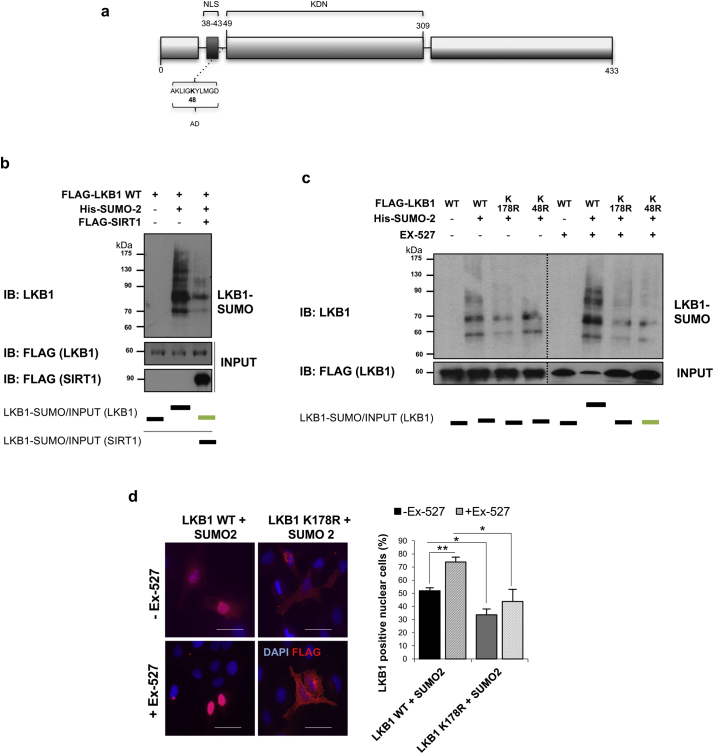
Fig. 5.
Liver Kinase B1 (LKB1) is modified by SUMO-2 in Lys178 after its acetylation at Lys48 in human hepatoma cells. a. Schematic representation of LKB1 showing the Nuclear (NLS) localization, acetylation domain (AD) and the Kinase domain (KDN). Acetylation LKB1 mutant used is shown. b. Ni2+-NTA agarose bead pulldown in Huh-7 human hepatoma cells after transfection with the pcDNA3-FLAG-LKB1 Wild type plasmid (LKB1 WT), His-SUMO-2 and treatment with sirtuin 1 (SIRT1). c. Ni2+-NTA agarose bead pulldown in Huh-7 human hepatoma cells after transfection the LKB1 WT, LKB1 acetylation mutant, LKB1 K48R or the LKB1 SUMOylation mutant, LKB1 K178R, with His-SUMO-2, in the presence and absence of the Ex-527, the SIRT1 inhibitor. Normalized quantifications relative to inputs are shown below each panel; d. Representative immunofluorescence staining for FLAG and quantifications in Huh-7 hepatoma cells after transfection with the LKB1 WT or the LKB1 SUMOylation mutant LKB1 K178R and SUMO-2 in the presence and absence of Ex-527. Scale bar corresponds to 50 μm. At least triplicates were used per experimental condition. Data is shown as mean ± SEM. *p < 0·05 and **p < 0·01 are indicated (Mann-Whitney U test).