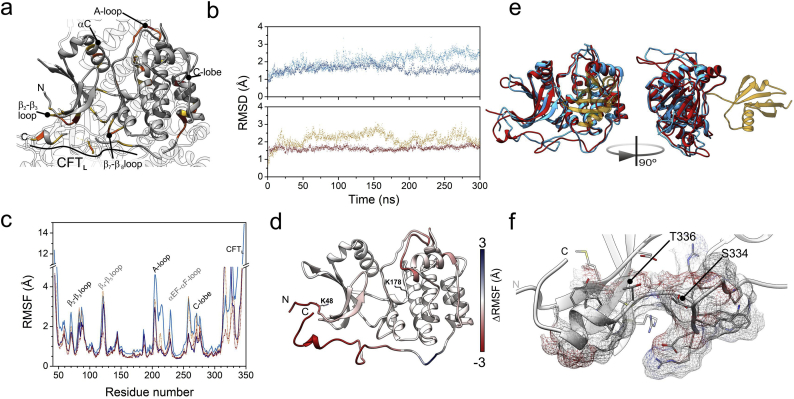
Fig. 6.
Model of structural and dynamic changes on Liver Kinase B1 (LKB1) upon post-translational changes. a. Structure of LKB1 in the context of its ternary complex with STRADα and MO25 [80] (pdb code: 2WTJ). Residues of LKB1 closer within 6 Å of either STRADα or Mo25 are in yellow, those closer than 4 Å are in orange. MO25 interacts with LKB1 through the A-loop, while STRADα interacts with the β2-β3 loop and the CFTL region. b. Comparison of the structures along the molecular dynamics computations. The Root Mean Square Deviation of trajectory snapshots upon alignment to the initial, energy minimized structure. Upper: data corresponding to unmodified LKB1 is in dark cyan, and that of K178-SUMOylated LKB1 is in blue. Lower: the trajectory of K48-acetylated LKB1 is in ochre, and dark red dots represent that computed for the K48-acetylated and K178-sumoylated. c. Per-residue atomic fluctuations computed along the last 250 ns of the trajectories. Colour attributes in b apply also to this panel. d. Map of changes in atomic fluctuations between unmodified LKB1 and fully modified (ALY at position 48, SUMO at position 178) onto the model obtained by simulated annealing of the mobile parts of LKB1 using the 2WTJ coordinates. Residues in red are more mobile in the unmodified protein, those in blue are more mobile in the fully modified one. e. Overlay of the structures of unmodified (cyan) and fully modified (red; SUMO in yellow) LKB1. Ribbons represent the coordinates closest to averages of the analyzed trajectory intervals. f. Detail of the terminal part of the CFTL region of the acetylated and SUMOylated STK domain of LKB1, according to the structure closest to the average of the last 250 ns of MD computations. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)