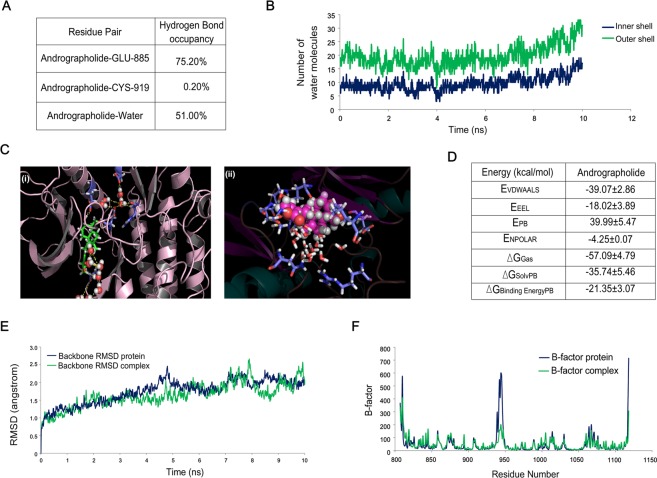
Figure 2.
Molecular dynamic simulation and docking study confirms that VEGFR2-andrographolide association is stable under physiological condition. (A) Hydrogen bond occupancy of andrographolide with Glu-885 and Cys-919 amino acid present in the VEGFR2 kinase domain. Andrographolide interaction with hydrogen of surrounding water molecules has also been represented here. (B) The graph represents inner shell of water molecules which is at a 3.4Å distance and second solvation shell is at a 5Å distance from the ligand. (C) Ribbon model of VEGFR2, stick form is andrographolide and sphere models are water molecules. Crystalline water molecules were kept intact during the molecular docking andrographolide, docked conformation suggests that water-mediated interactions are assisting binding of andrographolide. (D) Binding free energy component of andrographolide with VEGFR2 based on MMPBSA calculations. Abbreviations mentioned above denotes following- EVDWAALS: Non-Bonded van der Waals Energy. EEEL: Non-Bonded Electrostatic Energy. EPB: Polar solvation energy (PB). ENPOLAR: Nonpolar solvation energy from repulsive solute-solvent interactions (PB). ∆GGas: EVDWAALS + EEEL + Internal Energy. ∆GSolv: Polar solvation energy + Non polar solvation energy. ∆GBinding Energy: Binding Free Energy (∆GGas + ∆GSolv). (E) Root mean square displacement (RMSD) of VEGFR2 (protein) and VEGFR2-andrographolide. (F) B-factor analysis suggests flexibility of protein decreases after interaction with andrographolide.