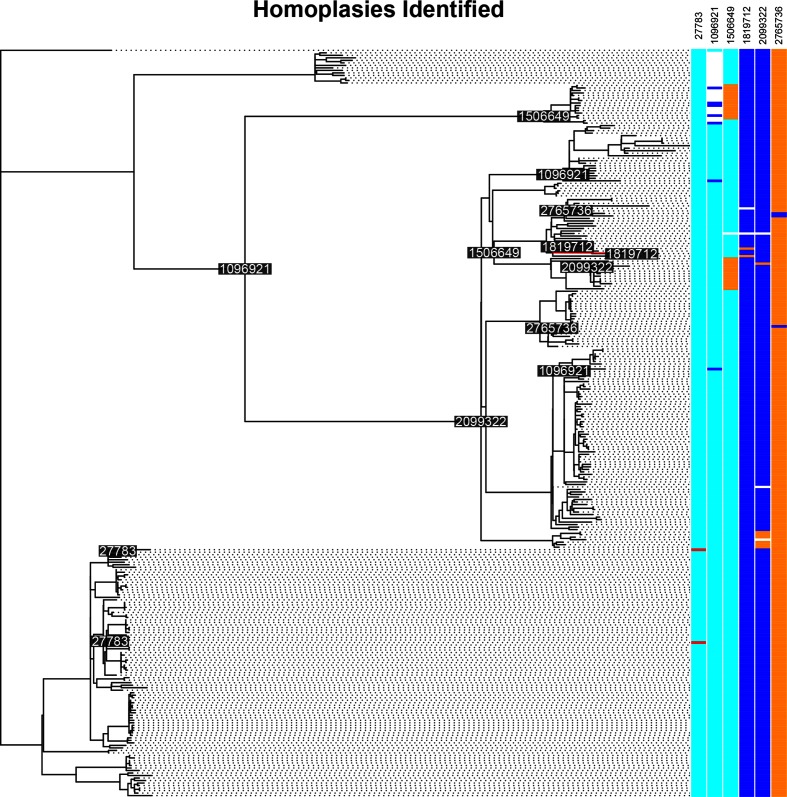
Fig. 5.
Identifying homoplasies in M. bovis data using HomoplasyFinder. A phylogenetic tree reconstructed using 298 published M. bovis whole genome sequences [25]. HomoplasyFinder identified homoplasies at six different positions (0.2 % of the 3852 polymorphic positions identified), the nucleotides associated with these positions in each sequence are plotted and coloured according to their type (Adenine=red, Cytosine=blue, Guanine=cyan and Thymine=orange). Where no information for the nucleotide at a particular site in a sequence was available, it is coloured white. The positions, on the M. bovis reference genome [33], associated with the identified homoplasies are reported in the top right and annotated on the internal nodes where a change was necessary. To avoid overlapping one of the labels was slightly moved and a red line points to the node it annotates.