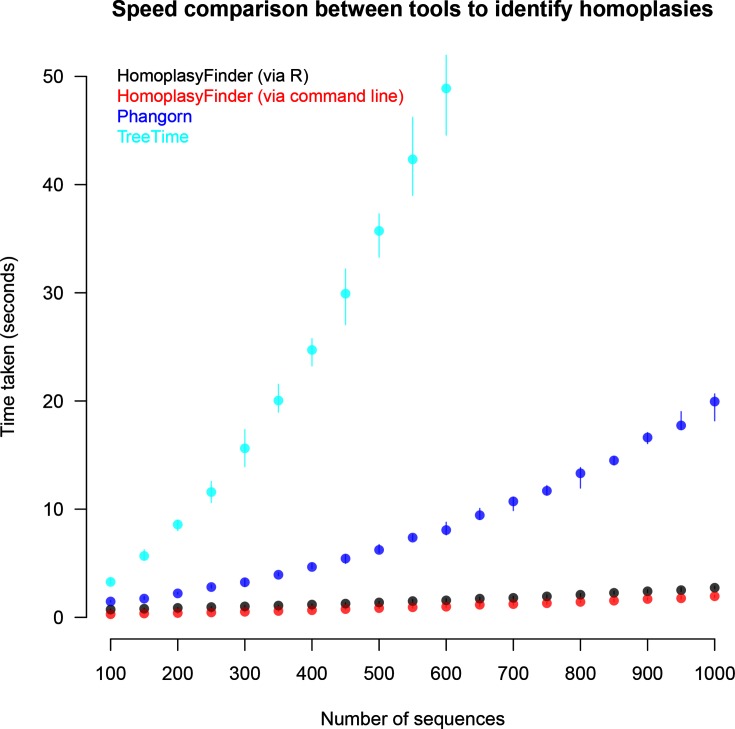
Fig. 6.
Comparing HomoplasyFinder to phangorn and treetime. The time taken to identify 10 homoplasies present in simulated phylogenetic datasets by HomoplasyFinder accessed in R and the command line, phangorn in R and by treetime in the command line. A total of 190 different datasets were tested, ranging from 100 to 1000 sequences, in steps of 50 with 10 replicates of each. The number of positions in these sequences ranged from 4000 to 20 000. The points and vertical lines plotted represent the mean, and range, respectively, of the ten replicates.