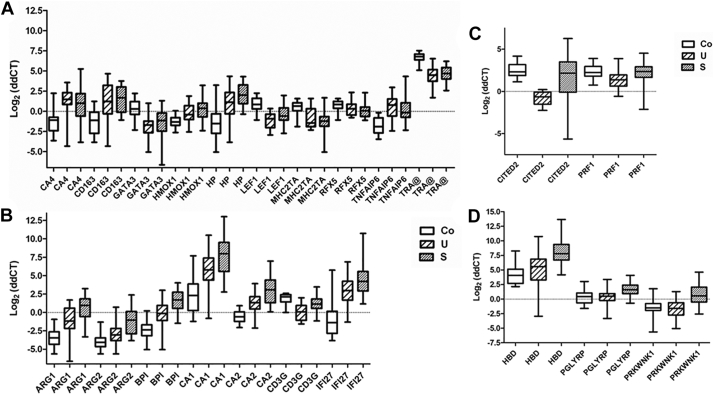
Fig. 4.
Expression pattern of selected genes validated by real-time RT-PCR. The expression pattern of selected genes was verified by Taqman real-time RT-PCR. Box-plots illustrate medians with 25 and 75 percentiles and whiskers from minimum to maximum. Groups were compared with the unpaired student's t-test and the P values corrected with the Bonferroni method. Genes are listed on the X-axis in alphabetical order. Co = controls; U = uncomplicated malaria; S = malaria associated with severe anemia. Genes similarly regulated in symptomatic malaria (A) include: CA4 (carbonic anhydrase 4), CD163 (CD163 scavenger receptor), GATA3 (GATA binding protein 3), HMOX1 (heme oxygenase 1), HP (haptoglobin), LEF1 (lymphoid enhancer-binding factor 1), MHC2TA (major histocompatibility complex class 2, transactivator), RFX5 (regulatory factor X 5), TNFAIP6 (tumor necrosis factor alpha induced protein 6), TRA@ (T cell receptor alpha). P < .0001 for UxCo, except for HMOX1 (P < .001), HP (P < .01) and RFX5 (not significant, but P < .01 for SxCo). Differences between S and U were not significant. Genes with strong up-regulation (except CD3G) in severe malaria (B) include: ARG1 (arginase 1), ARG2 (arginase 2), BPI (bactericidal permeability increasing protein), CA1 (carbonic anhydrase1), CA2 (carbonic anhydrase 2), CD3G (CD3 gamma), IFI27 (interferon-alpha inducible protein 27). P < .0001 for UxCo, except for ARG2 (P < .01). P < .0001 for SxU except for ARG1 and BPI (P < .001); CA1, CA2 and IFI27 (P < .01). Genes with unique regulation in uncomplicated malaria (C) include: CITED2 (Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2), PRF1 (perforin 1). P < .0001 for U ≠ Co, except for PRF1 (P < .01). Differences between S and Co were not significant. Genes uniquely expressed in severe malaria (D) include: HBD (hemoglobin delta), PGLYRP (peptidoglycan recognition protein 1), PRKWNK1 (lysine-deficient protein kinase 1). P < .0001 for SxU, except for PGLYRP (P < .001). Differences between U and Co were not significant.