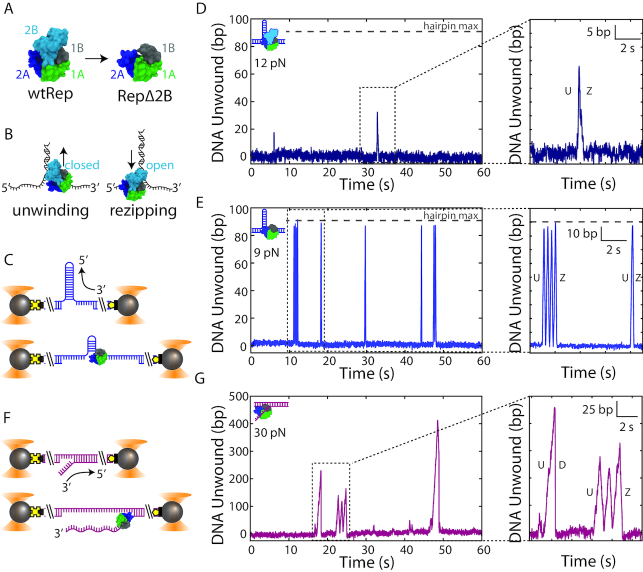
Figure 1.
Removal of the 2B subdomain activates Rep monomer unwinding. (A) wtRep structure in the ‘closed’ conformation (PDB 1uaa) showing its four subdomains (left), and a structural model of RepΔ2B with 2B replaced by three glycine residues (right). (B) Schematic of SF1 helicase (here, wtRep) monomer activity on DNA. 3′-to-5′ translocation on the 3′-strand of a DNA fork leads to unwinding of the duplex (left) with 2B in the closed state. 3′-to-5′ translocation on the 5′-strand leads to duplex rezipping behind the helicase (right) with 2B in the open state. (C) Schematic of dual-optical trap measurement of DNA hairpin unwinding. Two beads (grey spheres) held in traps (orange cones) stretch a DNA hairpin (blue) at constant tension via biotin-streptavidin (black square-yellow cross) and digoxigenin-anti-digoxigenin (yellow pentagon-black rectangle) linkages. Unwinding of a hairpin base pair releases two nucleotides, increasing DNA extension. (D) Representative data trace of DNA hairpin unwinding by wtRep dimer (schematic, top left corner). Close-up (inset) of one round of activity showing unwinding (U) followed by hairpin rezipping (Z). (E) Representative data trace of processive unwinding of DNA hairpin by RepΔ2B monomer (schematic, top left corner). The close-up (inset) of two rounds of activity shows hairpin unwinding (U) and rezipping (Z). The grey horizontal dashed line (D–E) indicates the limit expected for complete hairpin opening. (F) Schematic of dual-optical trap measurement of DNA fork (magenta) unwinding. Unwinding of a single base pair of the fork releases one nucleotide, changing DNA extension. (G) Representative data trace of processive DNA fork unwinding by RepΔ2B monomer (schematic, top left corner). The close-up (inset) shows two rounds of activity; protein unwinding (U) ends either by mid-fork dissociation (D) or dissociation at the base of the fork after three rounds of unwinding (U) and rezipping (Z).