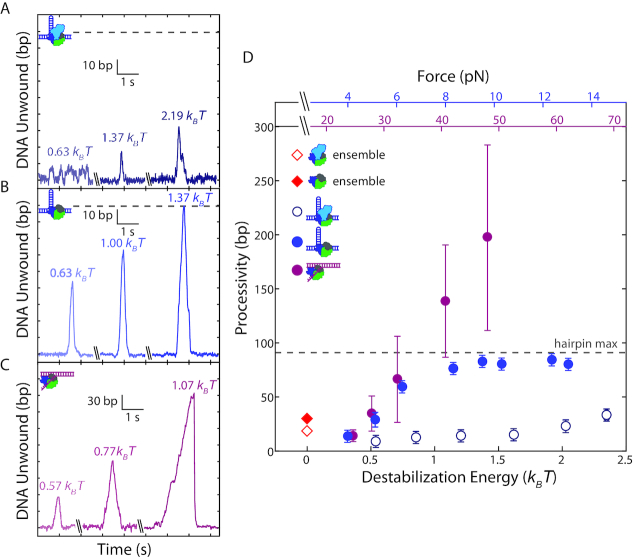
Figure 2.
Effect of duplex stability on unwinding processivity of RepΔ2B and wtRep. (A–C) Representative traces of hairpin DNA unwinding by wtRep dimer (A, shades of dark blue), hairpin DNA unwinding by RepΔ2B monomer (B, shades of blue), and fork DNA unwinding by RepΔ2B monomer (C, shades of magenta) showing unwinding processivity with increasing force. Values in units of kBT correspond to destabilization energies (see SI Text). (D) Median processivity as a function of destabilization energy for wtRep hairpin (open dark blue circles, N = 37–67), RepΔ2B hairpin (filled blue circles, N = 34–770), and RepΔ2B fork (filled magenta circles, N = 37–180) unwinding activity. Maximum processivity at zero force determined from stopped-flow ensemble measurements for wtRep dimer (red open diamond) and RepΔ2B monomer (red filled diamond). The top axes show the forces applied to the hairpin (blue) or fork (magenta), and the bottom axis shows the corresponding destabilization energy. Error bars represent standard error of the median. The gray dashed lines in A–B, D indicate the extension expected for complete hairpin opening, placing a limit on the maximum attainable processivity.