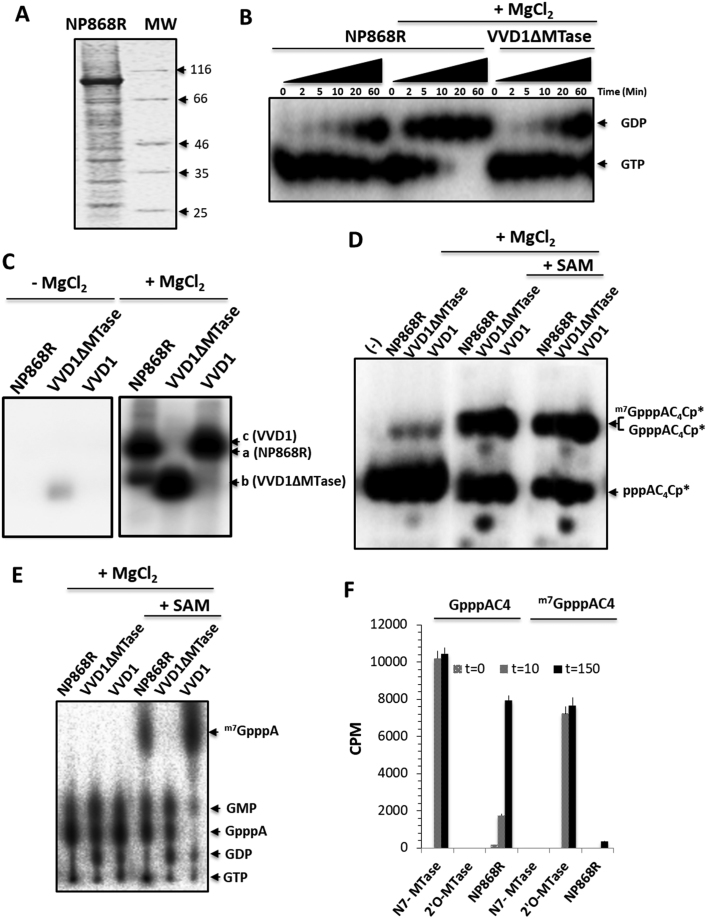
Figure 5.
NTPase, GTase and N7-MTase activities of NP868R capping enzyme. (A) Coomassie-stained, reducing SDS-PAGE showing the insect-cell derived NP868R protein purified by Ni2+-affinity chromatography migrating at around 100 kDa. Protein ladder is shown on the right lane. (B) NTPase activity. Autoradiographs of 20% urea-PAGE gel show that NP868R and the D1R subunit of VV (VVD1ΔMTase) hydrolyze [α-32P] radio-labeled GTP to GDP. The hydrolysis of GTP by NP868R is stimulated by 2.5 mM MgCl2. (C) GTP-binding assay. Autoradiographs of 12% SDS-PAGE show that NP868R, VVD1RΔMTase and VVD1R form covalent adducts, after 1 h of incubation with [α-32P] radio-labeled GTP in presence of 2.5 mM MgCl2. The main radiolabeled bands are detected at a molecular weight corresponding to the incubated proteins. (D) GTase activity. A synthetic RNA (pppAC4), radiolabeled at its 3′-end using [α-32P]-pCp (pppAC4-Cp*, Ctl) is incubated with NP868R, VVD1RΔMTase and VVD1R in presence of 1 mM GTP. The RNA reaction products, obtained after 1 h of incubation, are separated on a 20% urea–PAGE gel. The VV-capping enzyme (VVD1R and VVD1ΔMTase) used as positive control, convert pppAC4-Cp* into m7GpppAC4-Cp* or GpppAC4-Cp*, respectively. The autoradiographs show that, in presence of 2.5 mM MgCl2, NP868R convert pppAC4-Cp* RNA to a RNA migrating a molecular weight similar to the capped RNA generated by VVD1R proteins. (E) Cap structure identification. pppAC4 RNA was incubated with [α-32P]-GTP in presence of NP868R, VVD1RΔMTase and VVD1R respectively. The reaction products were digested by nuclease P1, and the caps were separated by thin-layer chromatography (TLC). m7GpppA is the predominant product seen in presence of the methyl-donor (SAM) using the VVD1, whereas VVD1RΔMTase synthesized mainly GpppA, as well as GTP hydrolysis products (GDP and GMP). m7GpppA cap structure is detected when NP868R is incubated with SAM and 2.5 mM MgCl2, and GpppA in absence of SAM. (F) MTase activity. Activity of NP868R was assessed by monitoring the transfer of tritiated methyl (CH3) from SAM to a short synthetic RNA (GpppAC4 or m7GpppAC4) after 10 and 150 min. The bar chart experiment compares the activities of NP868R with that of human N7-MTase (N7-MTase) and SARS-COV 2′O-MTase known to methylate specifically the N7 position of the RNA cap structure and the 2′O position of capped RNA respectively. NP868R methylate the GpppAC4 but not RNA already methylated on their N7 position (m7GpppAC4) suggesting that the methylation occurs on the N7 position of cap structure. The error bars indicate the standard deviation of three experiments.