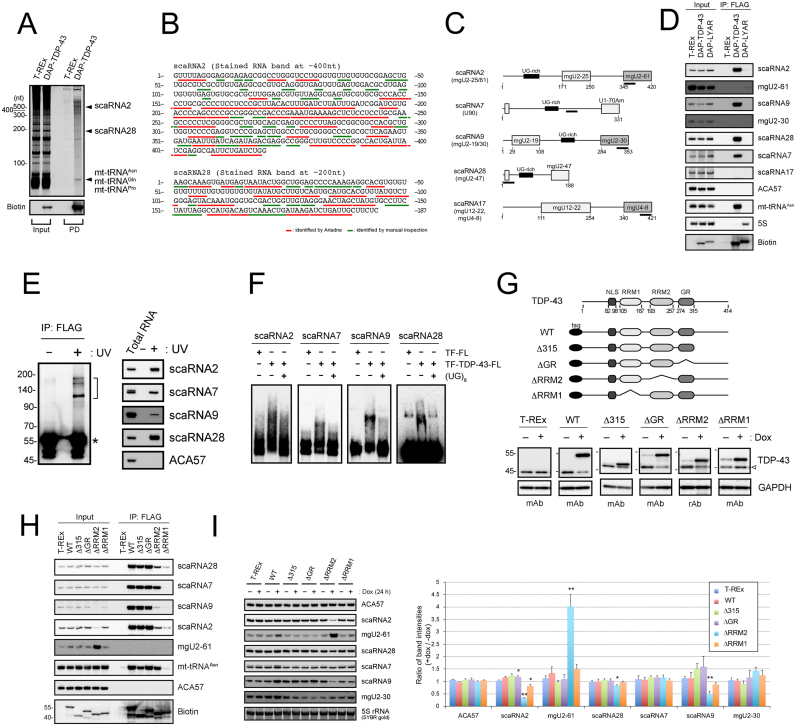
Figure 1.
TDP-43 binds C/D scaRNAs that have a UG-rich motif. (A) Isolation of RNAs bound to DAP-TDP-43 by two-step affinity purification (pull-down, PD; His-tag and FLAG-tag) from the cell extract of DAP-TDP-43-expressing T-REx 293 cells after a 24-h treatment with doxycycline. DAP-TDP-43 was detected by western blotting using horseradish peroxidase-conjugated streptavidin (streptavidin-HRP; Biotin). scaRNA2 and scaRNA28 were identified by in-gel RNase T1 digestion–LC-MS analysis. Input, total RNA (1 μg/cell line). (B) Oligonucleotides identified by LC-MS analysis of the in-gel RNase T1-digested RNA bands corresponding to scaRNA2 and scaRNA28 in Figure 1A are underlined in the sequence of scaRNA2 and scaRNA28. Red lines indicate oligonucleotides identified by Ariadne search, and green lines are by manual inspection. (C) Schematic structures of C/D scaRNAs (scaRNA2, 7, 9, 28 and 17). UG-rich motif and the guide domain for post-transcriptional modification are shown in each scaRNA. DNA probes used for northern blot analysis are indicated as black lines under each scaRNA. (D) DAP-TDP-43−binding RNAs immunoprecipitated using FLAG antibody (IP: FLAG) were detected by northern blot analysis with DNA probes hybridizing to the RNAs indicated on the right. The parental T-REx 293 cells and DAP-LYAR-expressing T-REx 293 cells were used as controls. DAP-TDP-43 and DAP-LYAR were detected by western blot analysis using streptavidin-HRP (Biotin). Input, total RNA (4 μg/cell line). (E) scaRNAs were isolated by the FLAG immunoprecipitation from DAP-TDP-43-expressing T-REx 293 cells with (+) or without (−) UV irradiation, separated by SDS-PGAE (left) and analyzed by northern blot analysis using DNA probes hybridizing to the RNAs indicated on the right (right). Half bracket indicates the position of the excised membrane. RNA-unbound DAP-TDP-43 detected by streptavidin-HRP is indicated by an asterisk. (F) Electrophoresis mobility shift assay using each of the biotin-labeled scaRNAs was done with (+) or without (−) recombinant TDP-43 (TF-TDP-43-FL), TF-FL and (UG)6 RNA oligonucleotide. (G) Schematic structures of DAP-TDP-43 deletion mutants (top). T-REx 293 cells or T-REx 293 cells expressing DAP-TDP-43 (WT) or its deletion mutants (Δ315, ΔGR, ΔRRM2 and ΔRRM1) were treated for 24 h with 100 ng/ml of doxycycline (dox), and the expression levels of DAP-TDP-43, its deletion mutants or endogenous TDP-43 were detected by western blot analysis with anti-TDP-43 antibodies (mouse monoclonal antibody, mAb; rabbit polyclonal antibody, rAb). GAPDH was used as a loading control. Open arrowhead shows endogenous TDP-43. (H) RNAs immunoprecipitated with DAP-TDP-43 (WT) and its deletion mutants (Δ315, ΔGR, ΔRRM1, ΔRRM2) using FLAG antibody (IP: FLAG) were detected by northern blot analysis with DNA probes hybridizing to the RNAs indicated on the right. The parental T-REx 293 cells were used as a control. DAP-TDP-43 and its deletion mutants were detected by western blot analysis using streptavidin-HRP (Biotin). Input, total RNA (4 μg/cell line). (I) T-REx 293 cells expressing TDP-43 (WT) or domain mutants (Δ315, ΔGR, ΔRRM2 and ΔRRM1) were treated with (+) or without (−) doxycycline for 24 h. Total RNAs were analyzed by northern blotting using DNA probes hybridizing to the RNAs indicated to the right. Ratios of staining band intensities (+dox/−dox) are shown by bar graph for the cells expressing TDP-43 or each of the domain mutants. Mean ± SEM, n = 3; *P < 0.05, **P < 0.01, unpaired t-test versus T-REx.