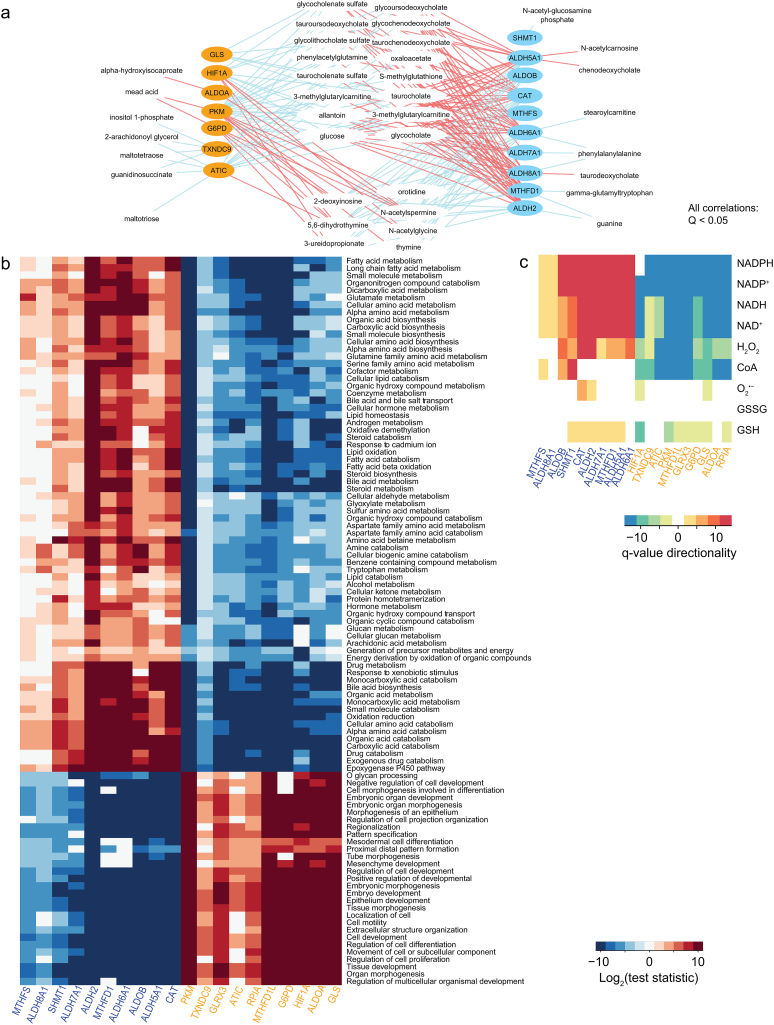
Fig. 3.
Opposite functional responses are observed in HCC tumors differentially expressing redox genes. A. Using metabolomics and gene expression data [14], we identified several metabolites with abundances that positively (red) or negatively (blue) correlated with the expression of genes in the ALDH2 and G6PD clusters (Q < 0.05, absolute Spearman's ρ > 0.4). Genes with expression that was not quantified by Ref. 14 are not indicated. B. GSEA for subjects expressing high and low redox genes in the ALDH2 (blue) and G6PD (orange) clusters. Only processes that were significant (Q < 0.05) in >4 columns are displayed (see Methods). Log2-normalized test statistics indicate the directionality of the processes (positive are upregulated, negative are downregulated, null are not statistically significant). C. Reporter metabolites were determined, and the minimum Q was computed for those with associated genes that were consistently up- or downregulated across compartments (Q < 0.05). Q values were normalized by computing −1 × log10(minimum Q), or log10(minimum Q), respectively, for up- or downregulated changes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)