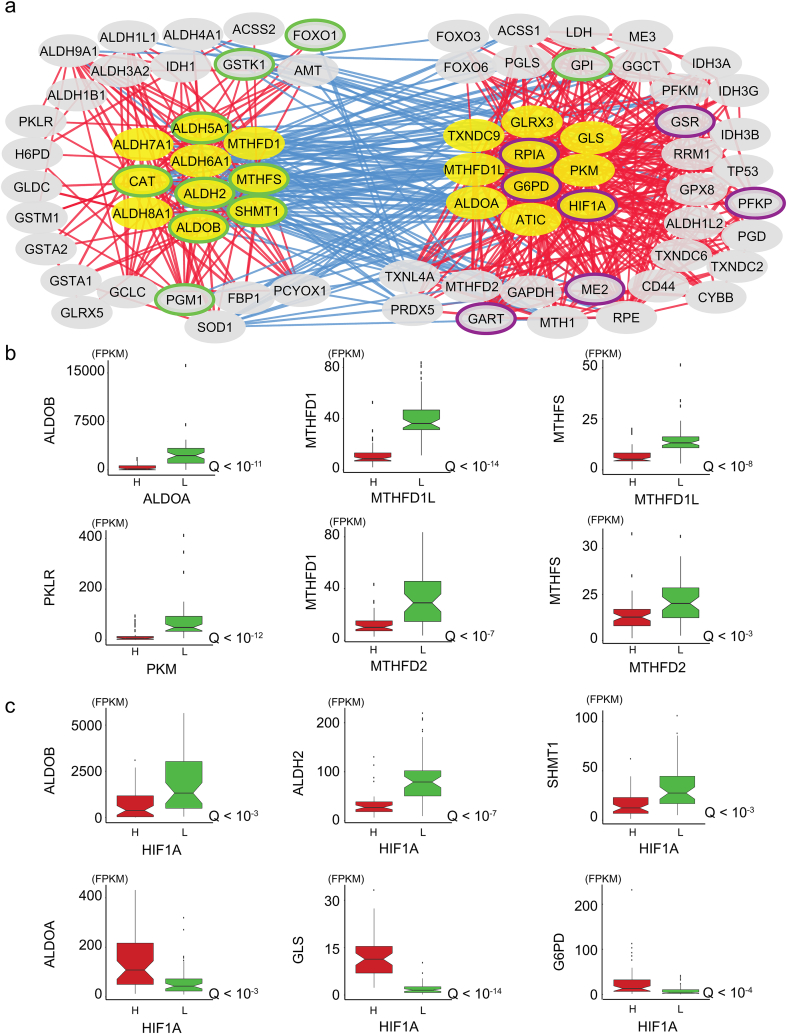
Fig. 4.
HCC tumors antagonistically co-express genes in ALDH2 and G6PD clusters, associated with opposite hypoxic environments. A. Genes in the two clusters (yellow) and their first neighbors (gray) are indicated. Redox genes that were positively co-expressed with genes in each cluster are displayed (absolute Spearman's ρ > 0.3, Q < 10−7). Green- and purple-circled nodes respectively indicate favorable and unfavorable prognostic genes (Table 1). Genes that were positively co-expressed with those in both clusters are not presented (e.g., NFE2L2, NNT, XCT, TXNRD1). Refer to Dataset 3 for the full list of correlations. B. Genes encoding enzyme isoforms or enzymes that catalyze alternative reactions in the same pathway were alternatively expressed. Boxplots display the expression of genes (FPKM) as a function of HCC samples displaying high (H) and low (L) expression of each gene (50 samples per group), and the respective Q values (Mann-Whitney U test). No significant difference (Q > 0.05) was observed for the other isoforms, with the exception of IDH1 as a function of IDH3A (Q < 0.005), for which high expression of the latter gene was associated with high expression of Idh1. Similar observations were obtained through differential expression analysis. C. Gene expression as a function of HIF1A expression. Note that among all genes in the ALDH2 and G6PD clusters, only the expression of MTHFD1, MTHFS and CAT did not significantly (Q < 0.05) differ between samples with high vs low HIF1A expression. For all boxplots, whiskers extended up to 1.5 times the interquartile range. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)