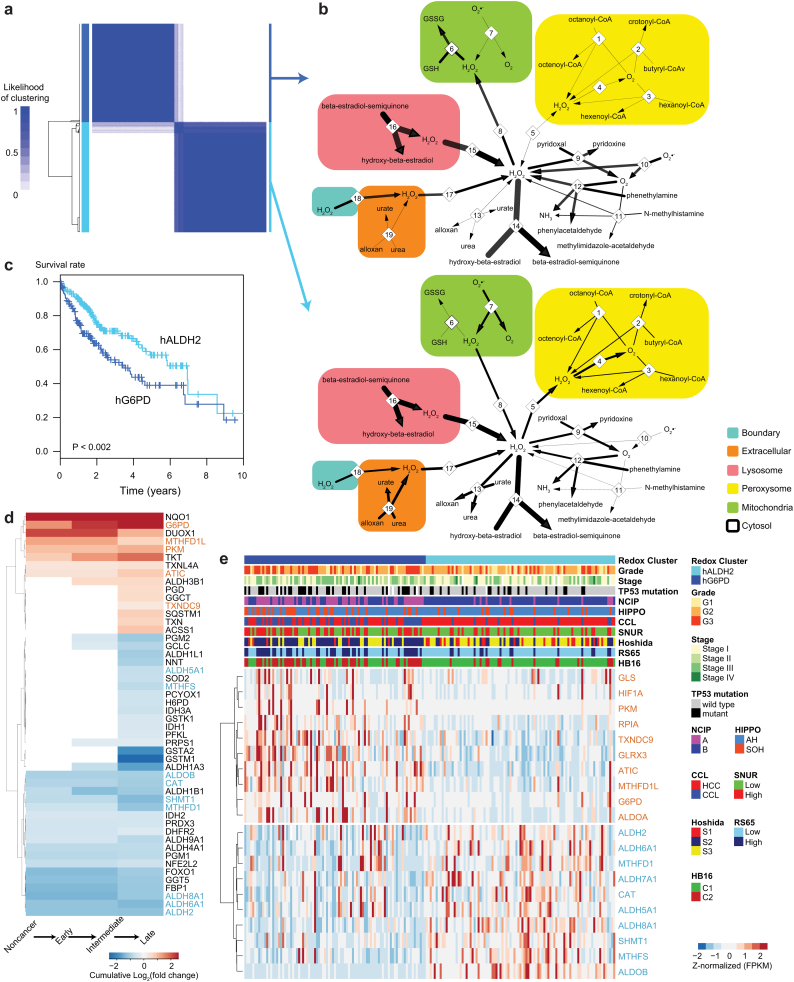
Fig. 5.
HCC tumors stratified based on redox genes display substantial differences in the survival and metabolism of H2O2, purines and fatty acids. A. Tumors were stratified based on the expression of genes in the ALDH2 and G6PD clusters through consensus clustering into two clusters that respectively displayed high expression of genes in ALDH2 and G6PD clusters (cyan and blue horizontal bars, respectively, hALDH2 and hG6PD). Clustering displayed the likelihood that a tumor was found in that cluster. Similar partitioning was achieved if all 174 redox genes were considered, reinforcing the robustness of the clustering. Note that >94% of the samples were consistently clustered in the two clusters (likelihood > 0.9). B. Metabolic differences displayed by the two groups of tumors as determined through metabolic modeling. GEMs for hG6PD (top) and hALDH2 (bottom) tumors were integrated with the gene expression data. Cell compartments are indicated by colored boxes. Numbers indicate enzyme-catalyzed reactions or transport reactions, as indicated in Dataset 11. Thin arrows indicate reactions with low or null fluxes (B: 6, 10, 11; C: 1–5, 7, 13, 19). Bidirectional reactions (e.g., transport reactions) indicate the estimated direction based on the flux value. Common metabolites (e.g., H2O and H+) are not displayed. C. Kaplan-Meier survival plot for hALDH2 (cyan) and hG6PD (blue) clusters indicates substantially lower survival for patients in the latter cluster (respective median 5-year survival of 0.57 and 0.39, P ≈ 0.0018). D. Differential expression analysis showing the opposing behavior between ALDH2-co-expressed and G6PD-co-expressed genes. Cumulative log2(fold changes) were computed for the significantly differentially expressed redox genes (Q < 0.05, DESeq2) between late (G3 + G4) vs intermediate (G2) vs early (G1) HCC and noncancerous samples. Genes in the ALDH2 (cyan) and G6PD (blue) clusters (Fig. 1B, inset). The full output is displayed in Dataset 15. E. Expression of genes in ALDH2 and G6PD clusters and detailed clinical information for 186 tumor samples (Cancer Genome Atlas Research Network, 2017). Gene expression data (FPKM) were row-normalized and clustered. Columns were sorted according to the redox cluster. Clinical information is shown for grade, stage, TP53 mutation status, and known tumor subtypes (NCIP, HIPPO, CCL, SNUR, Hoshida, RS65, HB16). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)